Overview
- Parallel processing, data binning for efficient computing
- Overfitting prevention: Regularization settings available
- Constructs level-wise trees.
Spliting Thresholds
- By default, XGBoost considers all the features for each split. However, by using hyperparameters such as colsample_bytree, colsample_bylevel, and colsample_bynode, it is possible to sample only a subset of features.
- When searching for a split point (threshold), instead of examining every possible value individually, the data is divided into several bins, and the optimal split point is determined. This method is called the Histogram-based Split Finding Algorithm.
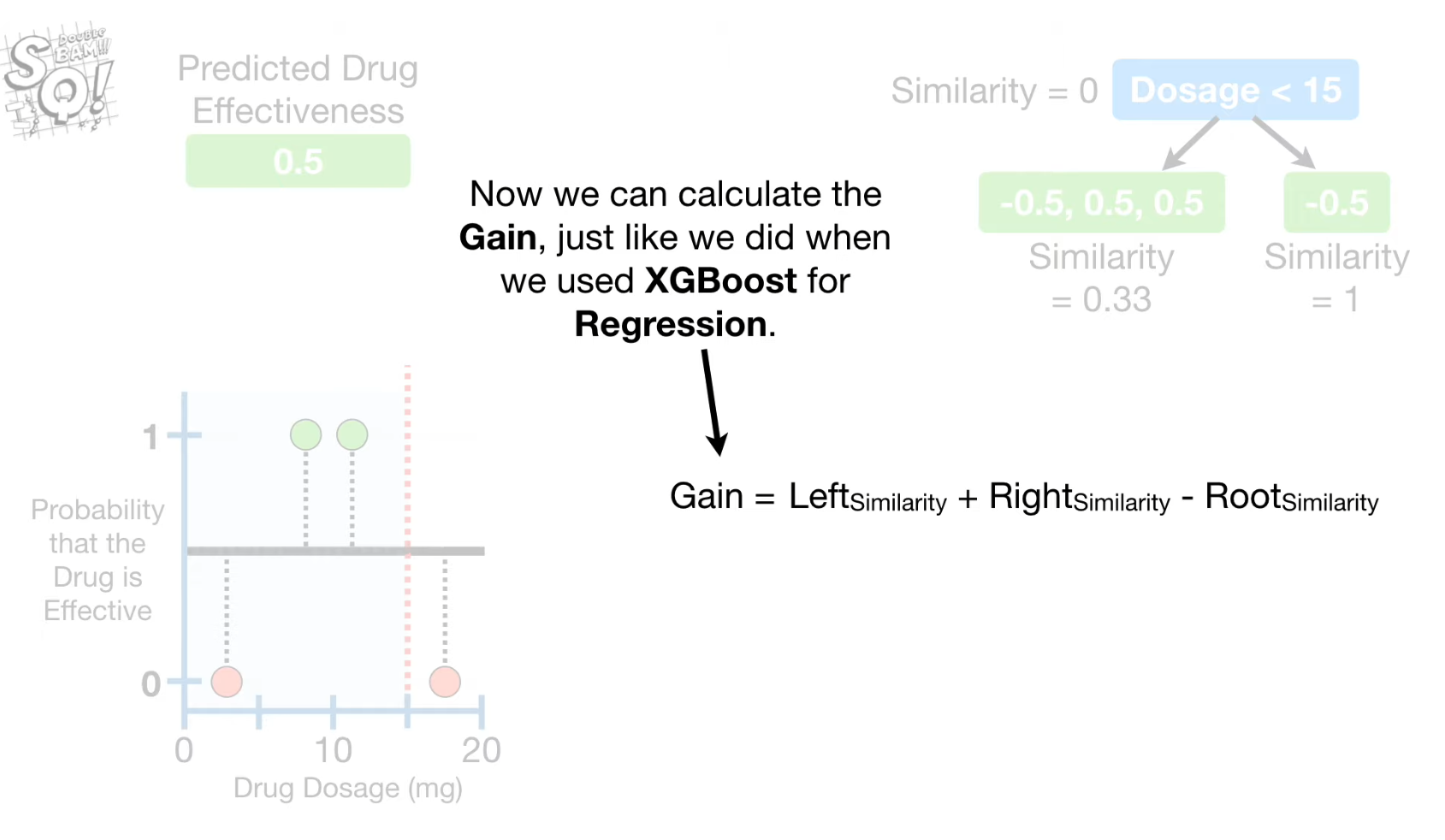
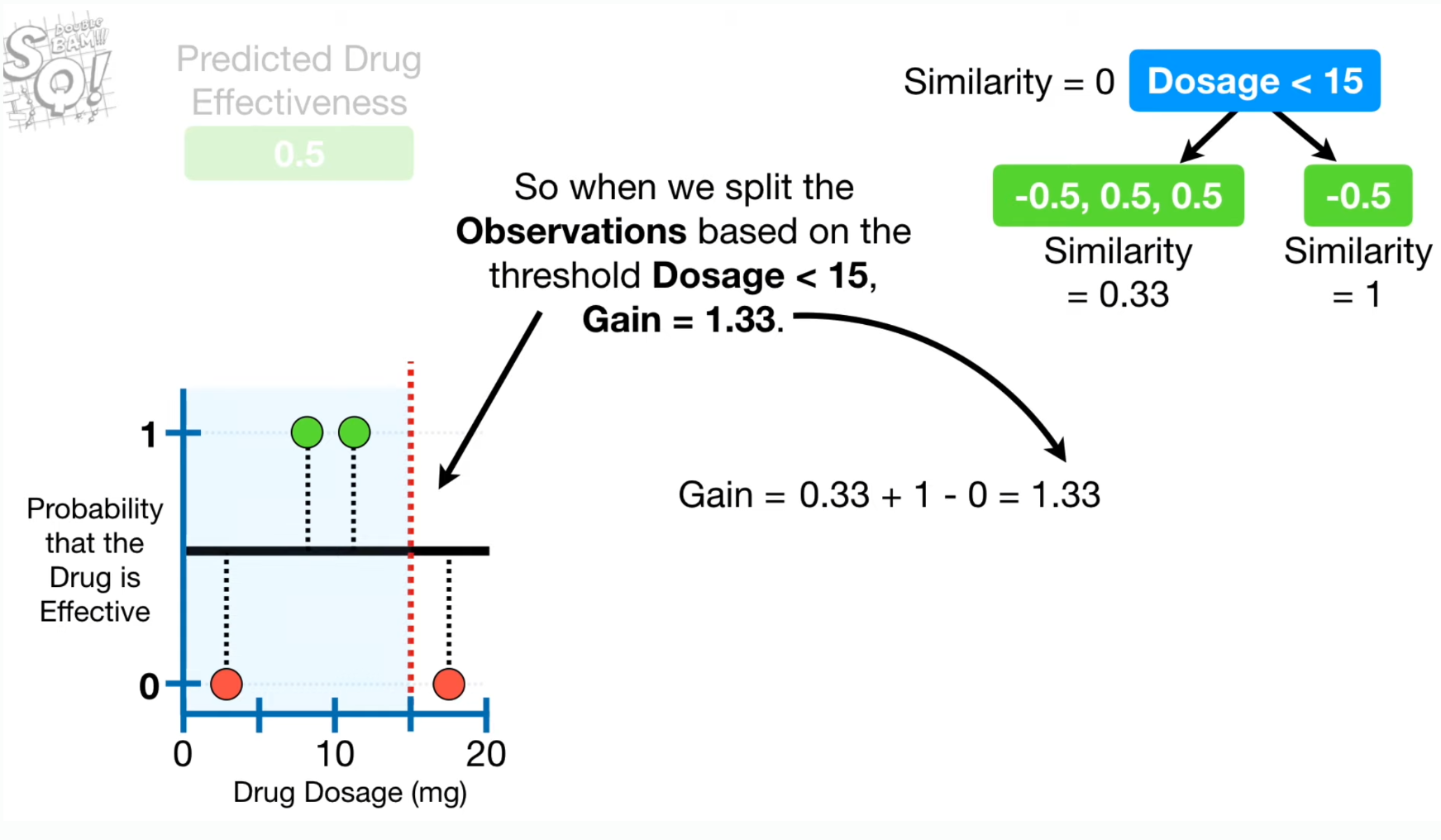
- Spliting point is decided based on Gain which is calculated from Similarity Scores at each node.
Steps
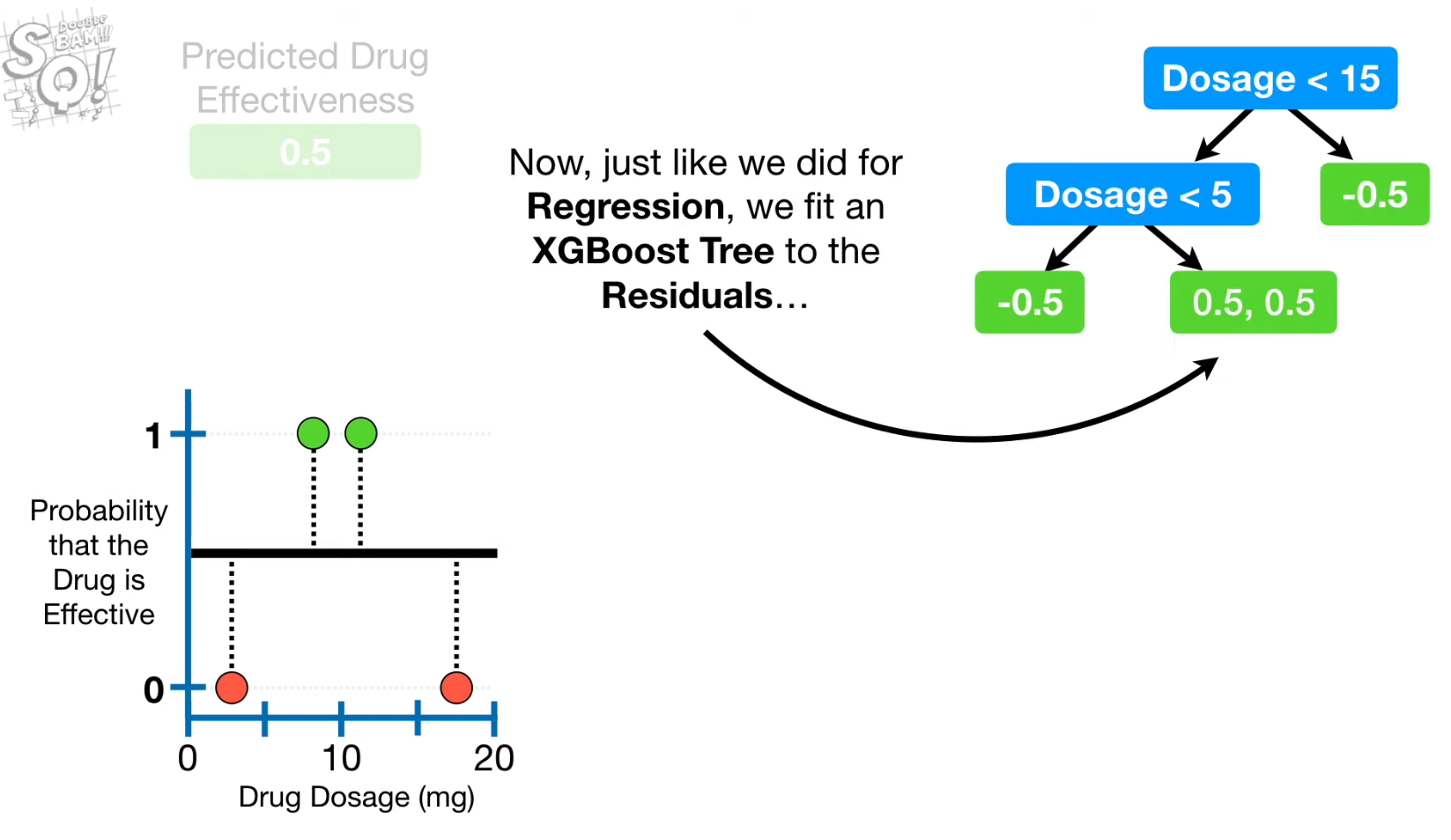
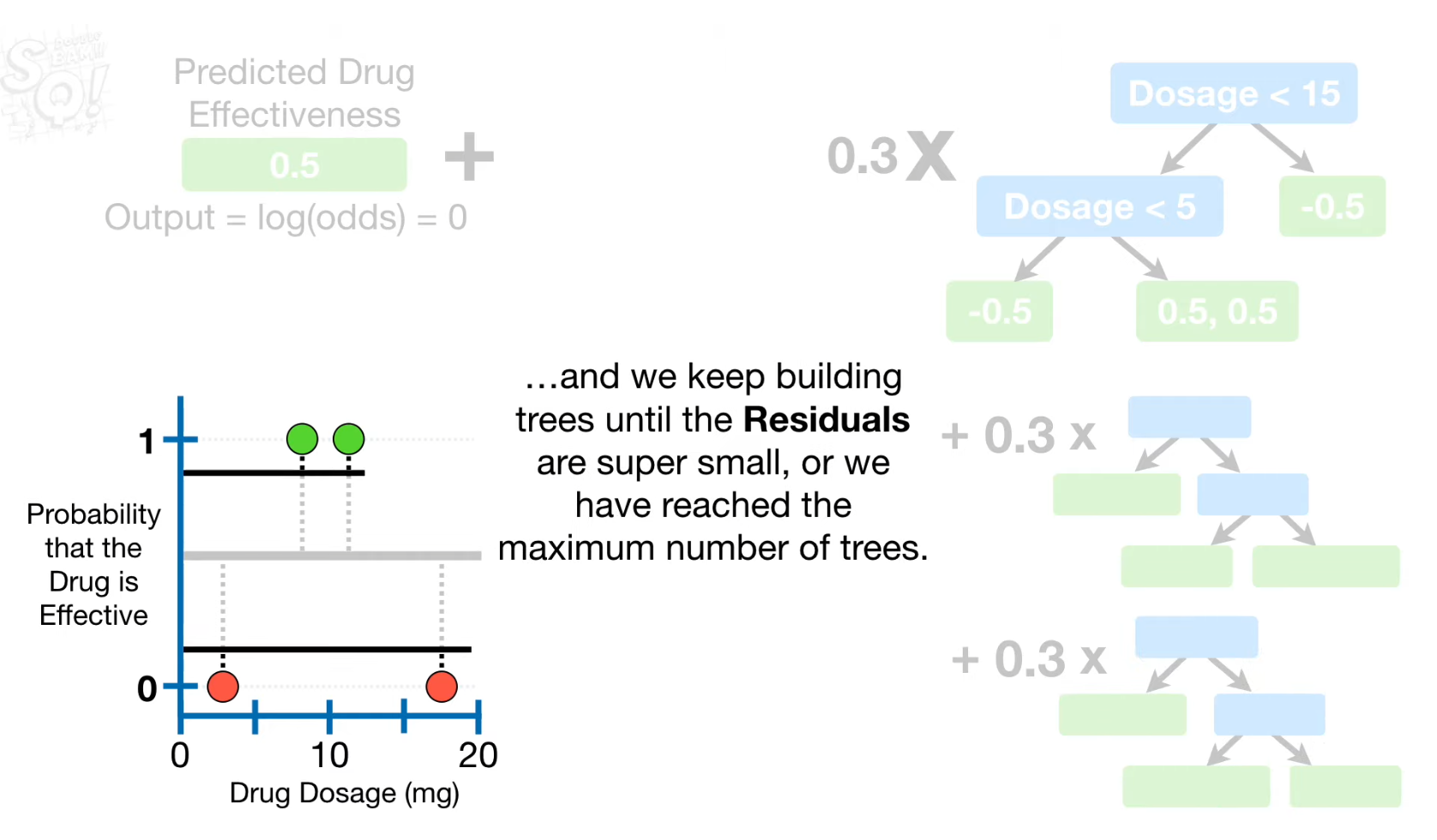
- Residuals 계산
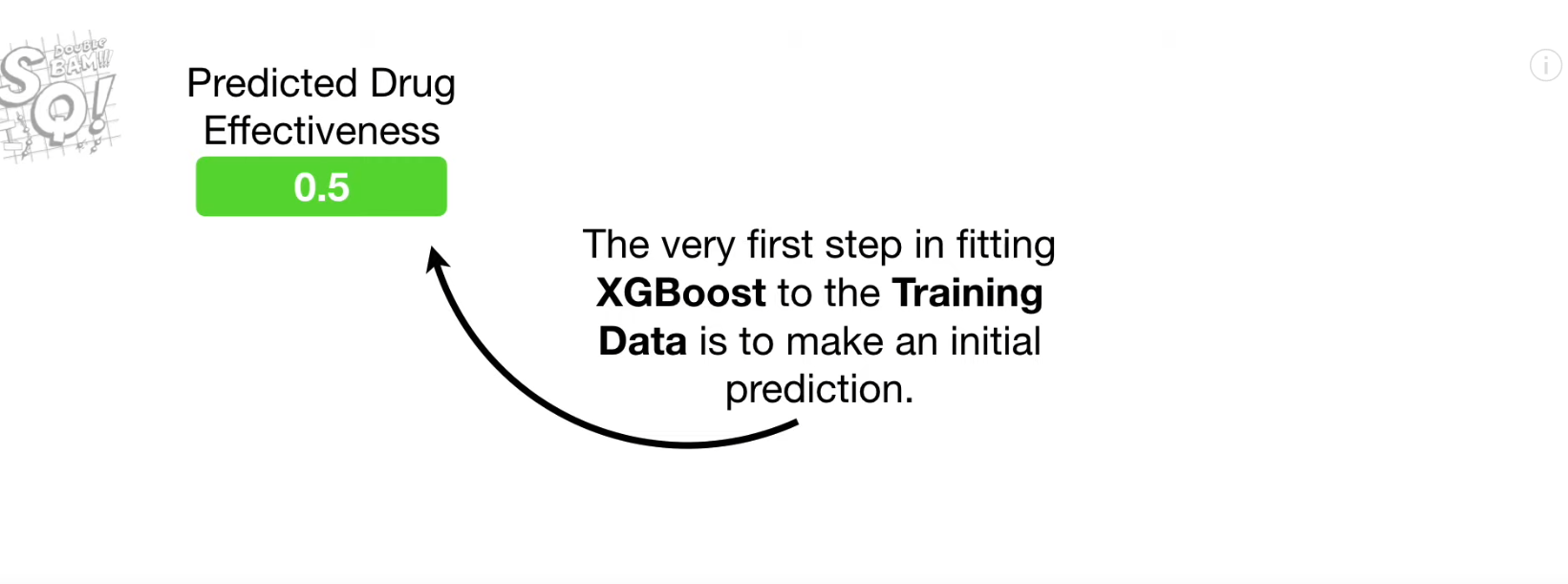
- Predicted value 계산 (initial default = 0.5)
- Split point 찾기
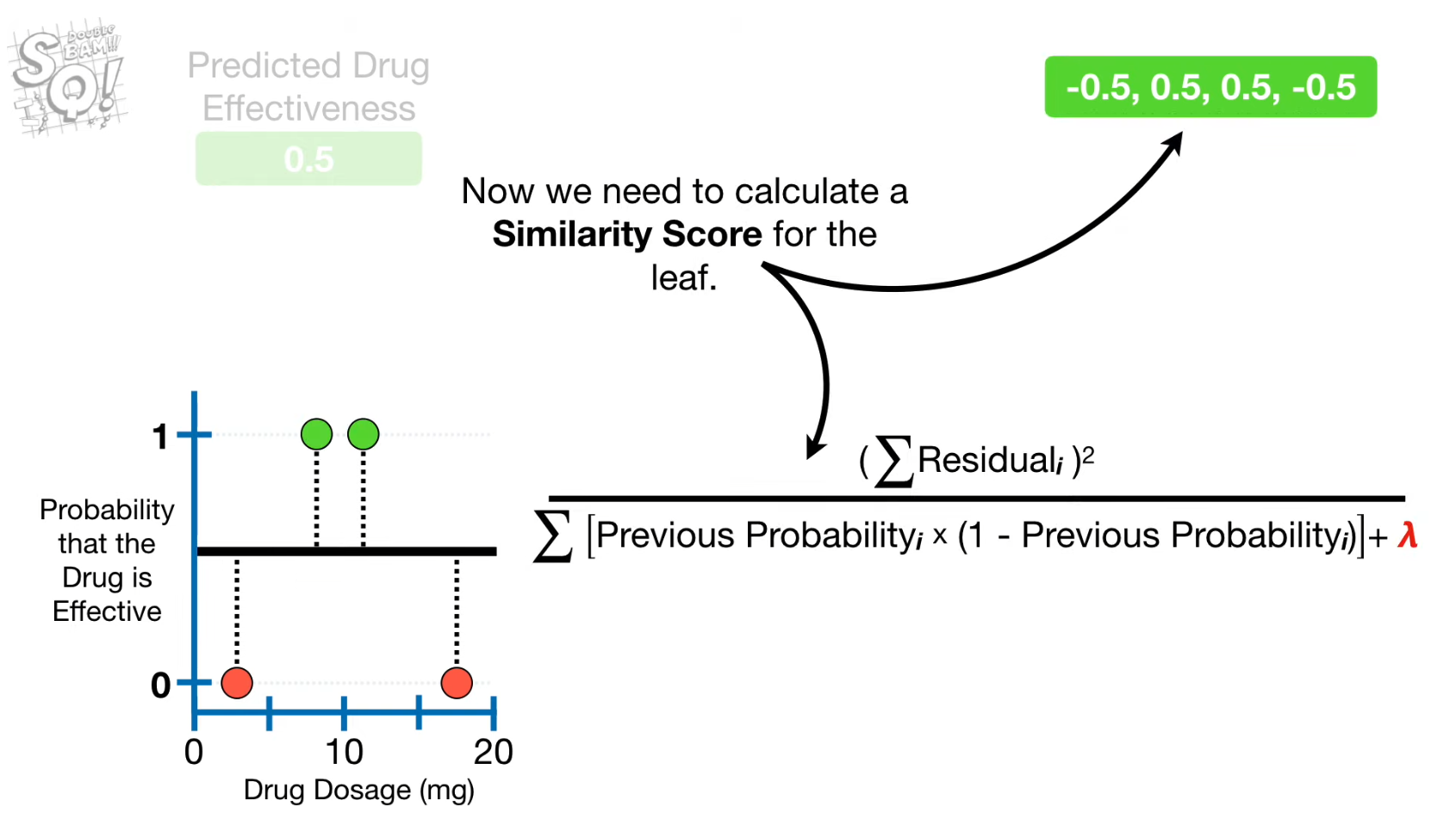
- "Similarity Score" for each node 계산
- "Gain" for each split using the similarity score 계산
- "Gain"이 가장 큰 feature와 threshold 계산
- Prune with "Gamma" (Gain - Gamma)
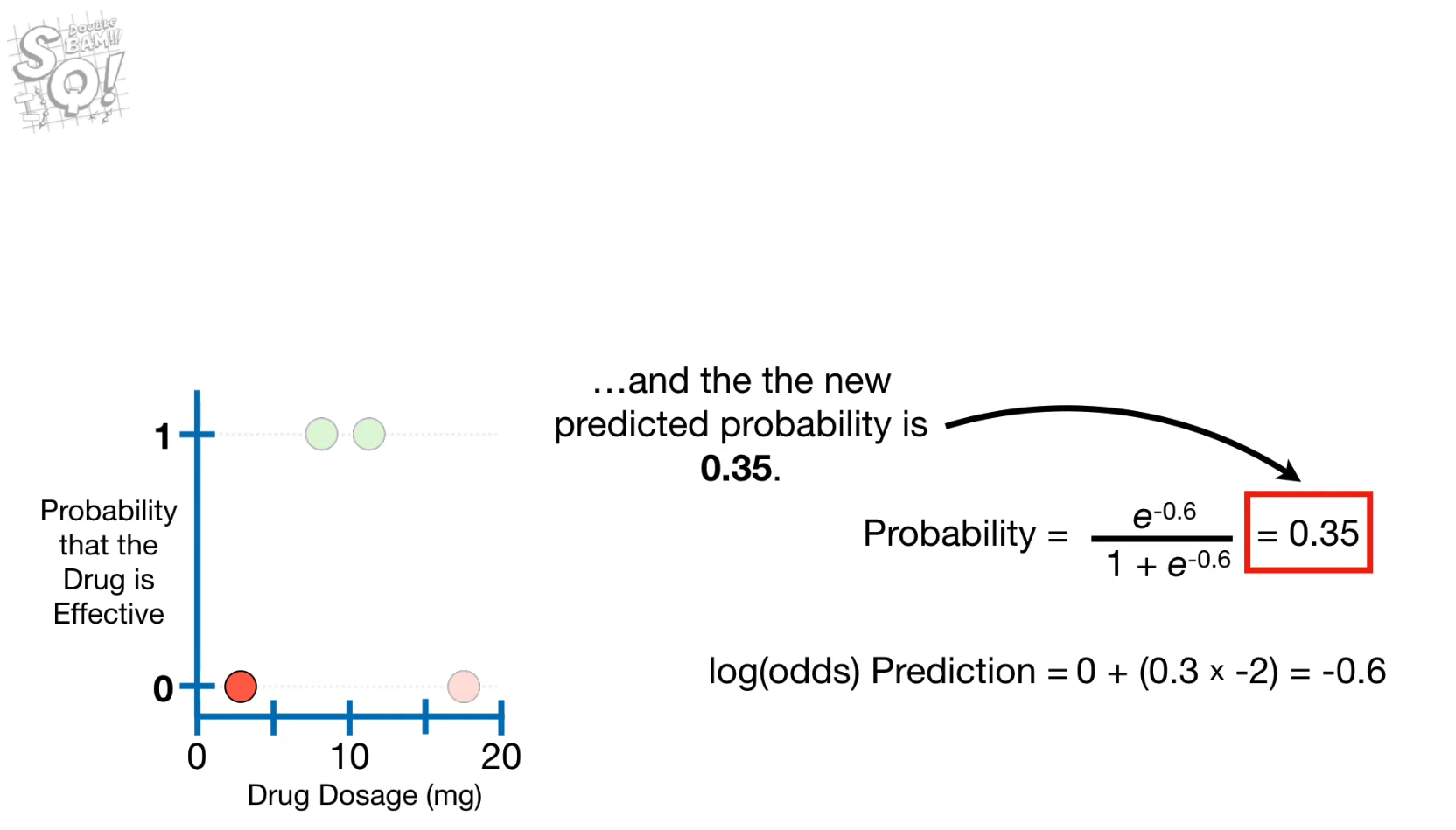
- Update the predicted probability for each observation
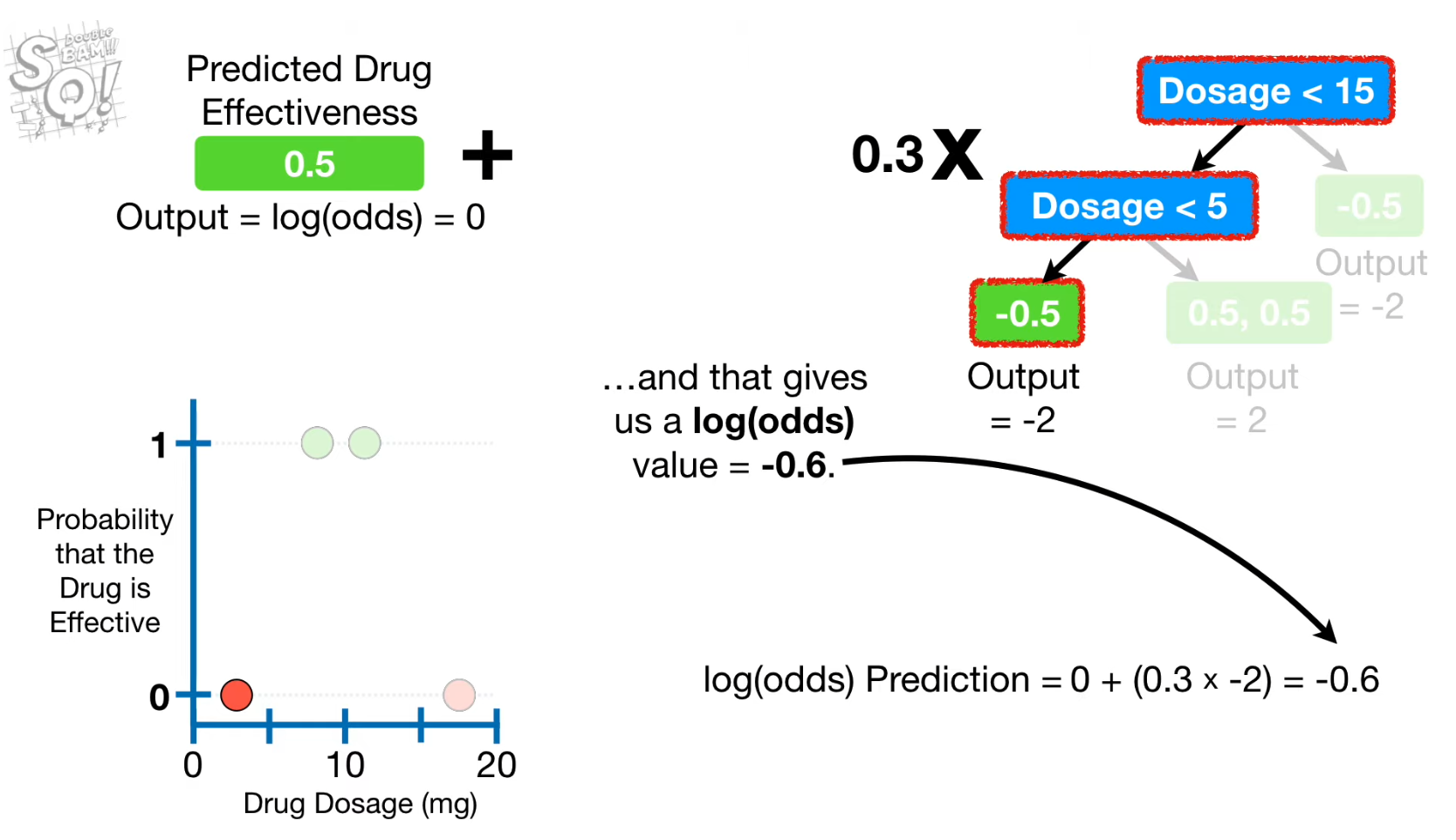
- Convert the previous predicted probability into an output value using log(odds)
- Update the output value for each node
- Convert the output value into the predicted probability using logistic function
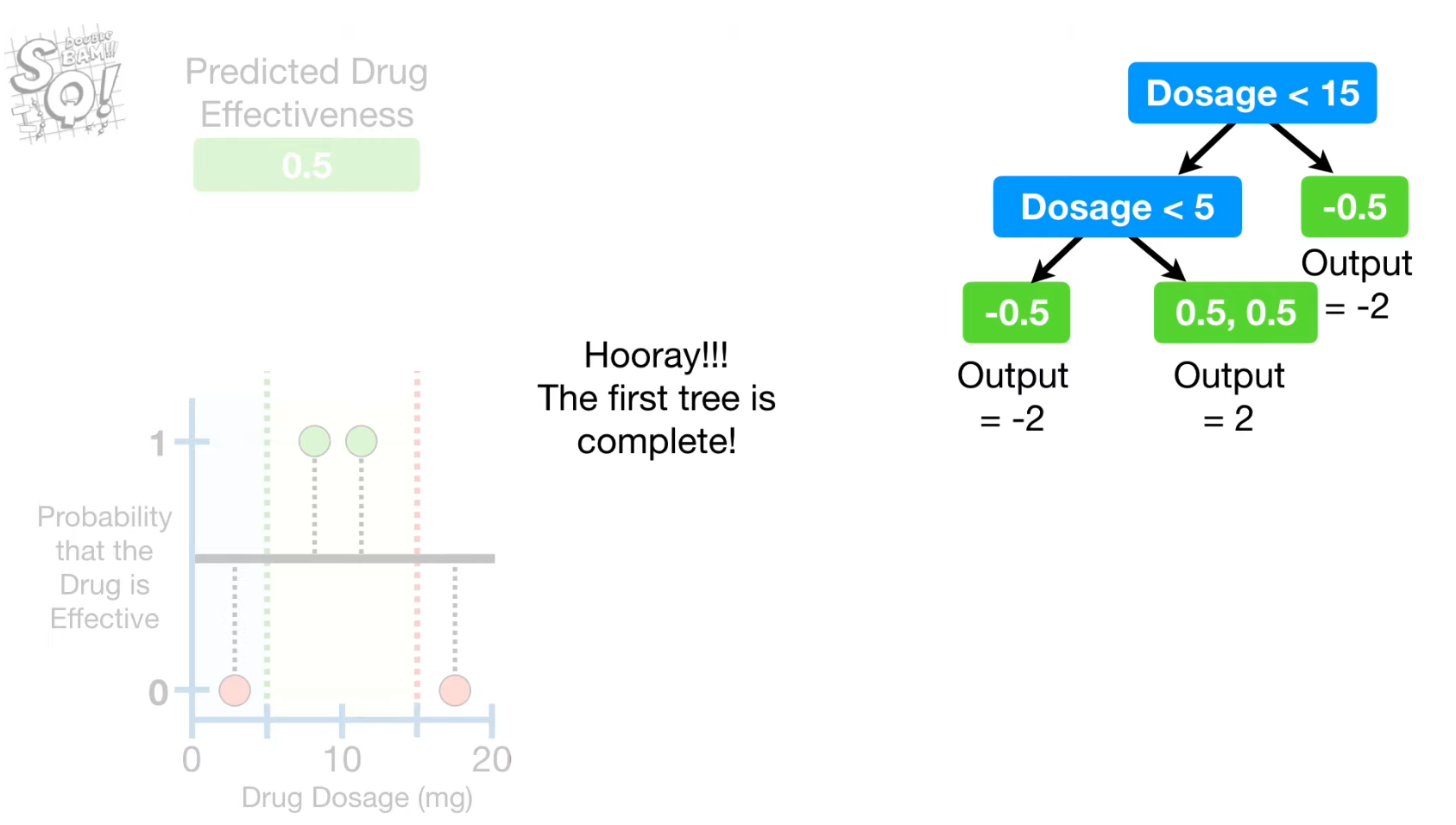
- Build the second tree using the new predicted probability
Hyperparameters
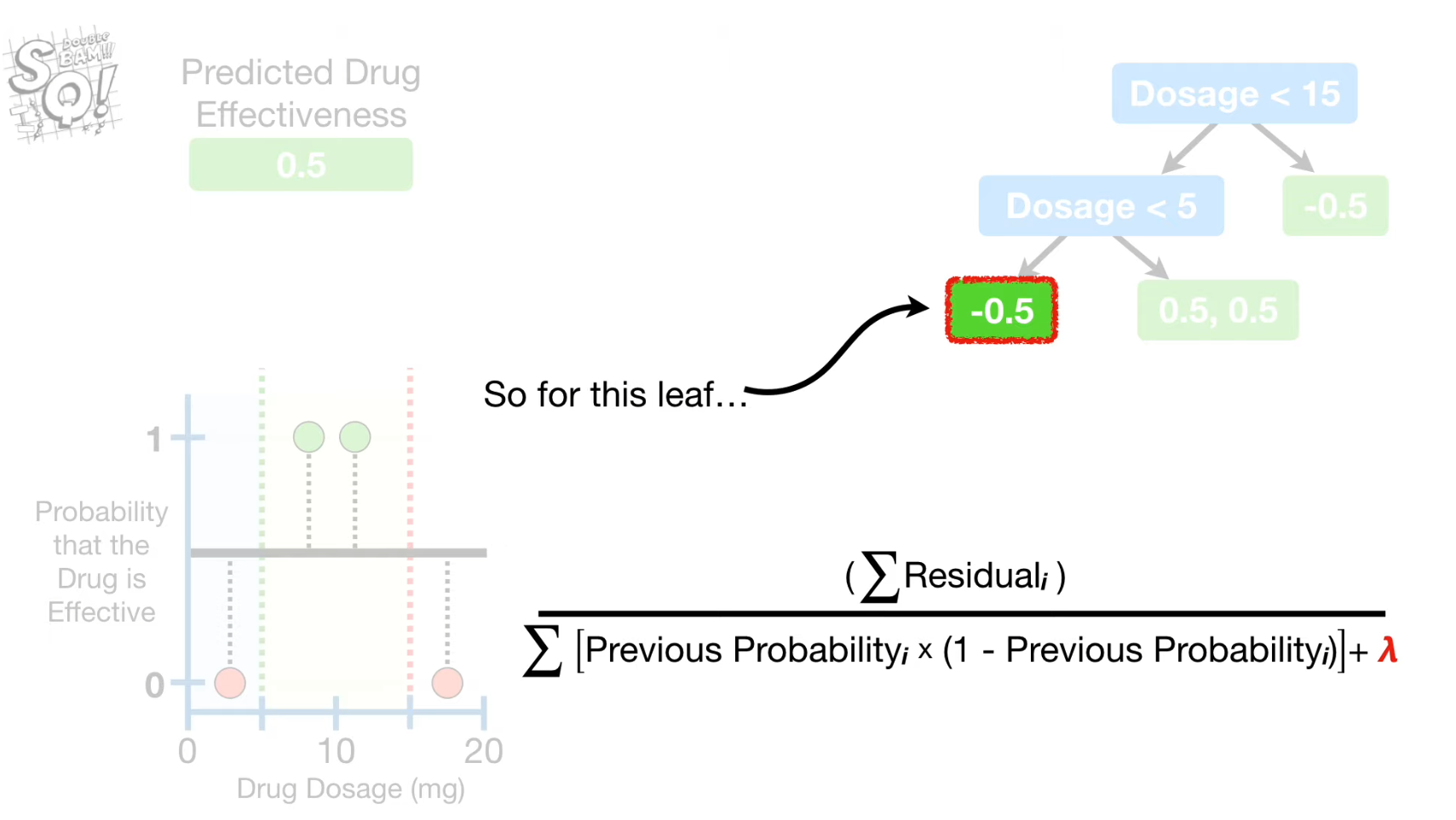
Lambda(Regularization parametre)
- reduces the similarity score → lower value of gain → prone to pruning with Gamma
- reduces the output value → lower predicted probability
Alpha(Learning rate)
- used to update the output value
Cover(min # residuals in a leaf)
Gamma
- if Gain - Gamma is negative, prune the parent node
Steps in detail