[Paper Review] Predicting survival from histology in colorectal cancer using deep learning
Paper Review
Predicting survival from histology in colorectal cancer using deep learning
Introduction
Colorectal cancer 환자의 HE stained tissue slide를 tile로 쪼개 9개의 tissue class로 segmentation, classification 을 진행하고, 환자의 overall survival과 관련된 5개의 stromal feature을 이용해 deep stroma score를 구함. 이를 internal test set, external test set 예후 예측에 넣어 OS 뿐만 아니라, relapse-free survival 에서 그리고 특정 tumor stage 내에서 independent prognostic factor임을 확인함.
Method
Patient cohorts and data availability
All images were 224 x 244 pixels and 0.5 um/px
1) 86 HE slides from NCT biobank and UMM pathology archive
NCT-CRC_HE-100K, 100,000 image patches
2) 25 HE slides from DACHS study in NCT biobank
CRC-VAL-HE-7K, 7,180 image patches
3)862 HE slides from 500 CRC patients from TCGA cohort
With matched RNA-seq data, calculated cancer-associated fibroblast CAF score
4) 409 HE slides from 409 patients in DACHS cohort in NCT biobank
With follow-up data, overall survival, diseases-specific survival and relapse-free survival
Training and testing of neural networks
CNN - five different CNN models (VGG19, AlexNet, SqueezeNet, GoogLeNet, Resnet50)
NCT-CRC-HE-100K : training(70%), validation(15%), testing(15%)
VGG19 had best performance (98.7 accuracy) -> further experiments 에서 사용
After neural network training with all 100,000 image patches, assessed tissue classification accuracy in external validation set: CRC-VAL-HE-7K dataset
Applied network to larger images with heterogeneous tissue composition
: sliding window to extract partially overlapping tiles, activations of softmax output layer saved.
For assessment of neural network training:
1) validation accuracy in independent training set
2) visualization with tSNE
3) DeepDream visualization
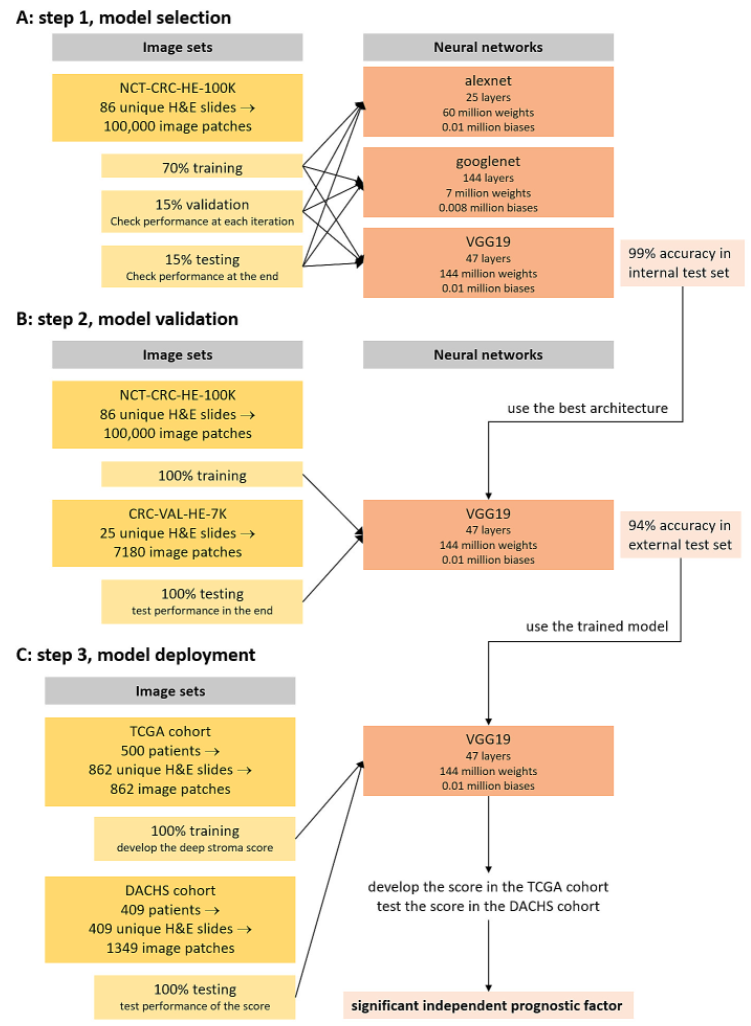
Deep stroma score (model deployment)
TCGA dataset의 image에서, mean activation of softmax output neuron for nine output classes in regions of 1500 x 1500 px (750 x 750 um)
Tissue decomposition of all images in TCGA set - assessed the prognostic performance of each tissue class (component) using univariable Cox PH model with continuous predictors.
ROC analysis with highest Youden index cutoff 로 optimal cutoff for survival prediction 설정. (확률 threshold)
HR (hazard ratio) > 1 인 tissue component의 정보를 combine 함.
Counted number of tissue classes (0 to 5) above Youden threshold for each class
Weight by the HR for each class (higher prognostic factor에 가중치 두기 위함.)
: deep stromal score
이후, DACHS external test set에서 score 상위 34% 는 high, 66%는 low value를 사용하여 sex, age in decades, UICC stage를 사용하여 multivariate cox ph model을 fit함.
Results
CNN performance for nine-tissue-components classification
VGG16 model: 100K tiles for train, 7K tiles for test
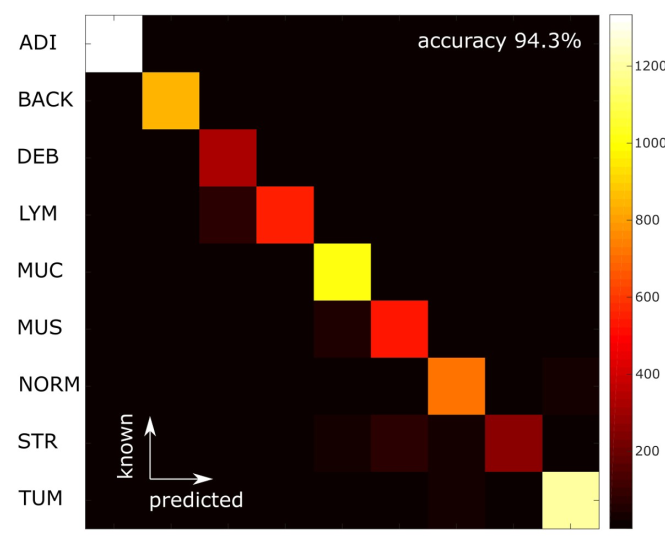
Misclassification은 muscle/stroma와 lymphocytes/debris 사이였고, 이 원인은 각각 비슷한 fibrous architecture, necrosis와 inflammatory cell의 상관관계라고 설명함.

tSNE를 사용해서 확인해본 결과 class 간 perfect 에 가까운 separation을 보여줌.
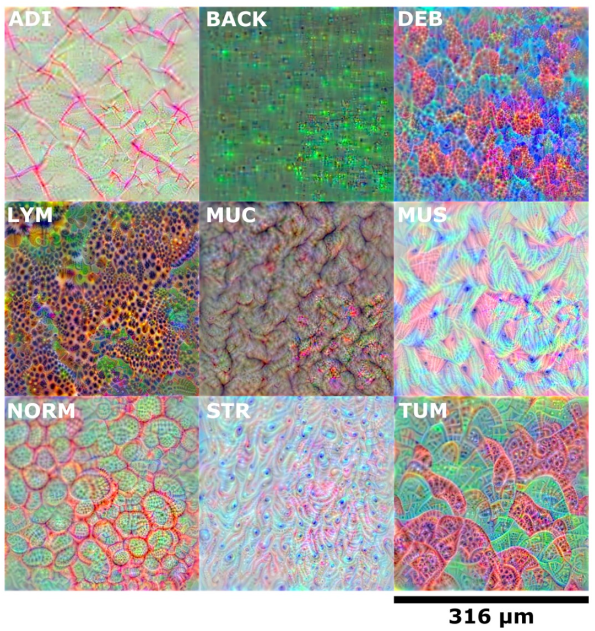
DeepDream approach를 사용해서, network가 학습한 morphological feature를 visuazlie한 결과 잘 학습한 것을 확인함.

larger image에 대해 neural network 를 apply함.

Tumor과 stroma class로 예측된 tiles에 대해 tSNE로 deep layer activation을 visualization함.
Stroma 내에서도 dense stroma, loose stroma가 분리되었고, tumor 내에서 poorly, well differentiated tumor region이 잘 분리되었음.
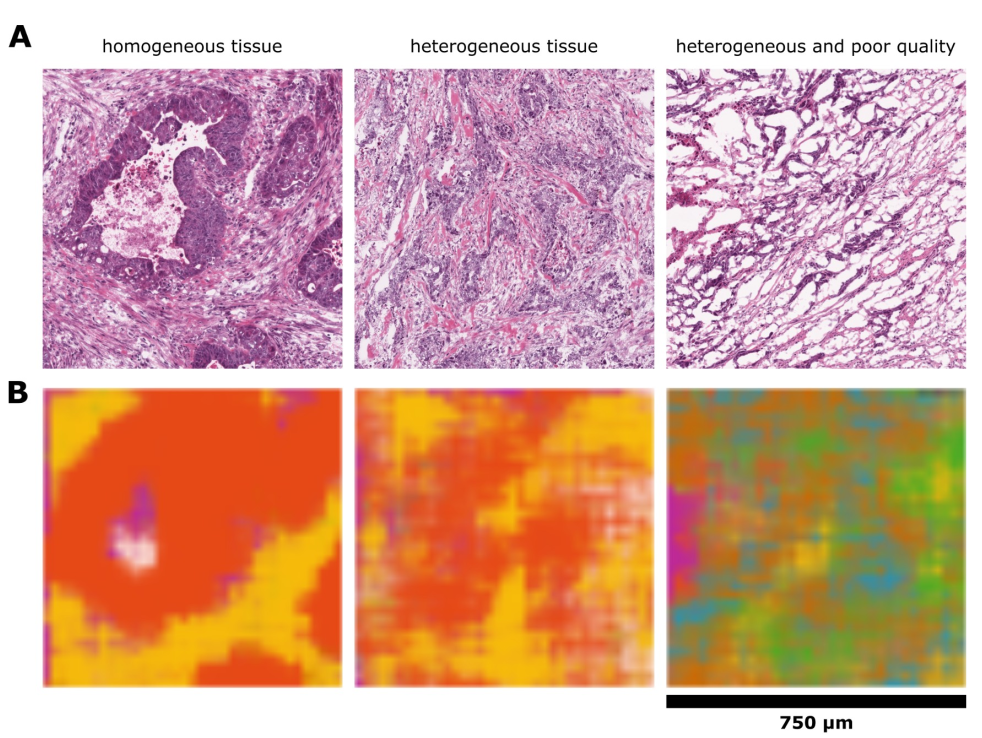
Varying quality를 가진 TCGA image에서 partially overlapping tile 방식으로 plausible neural network activation maps를 얻음.
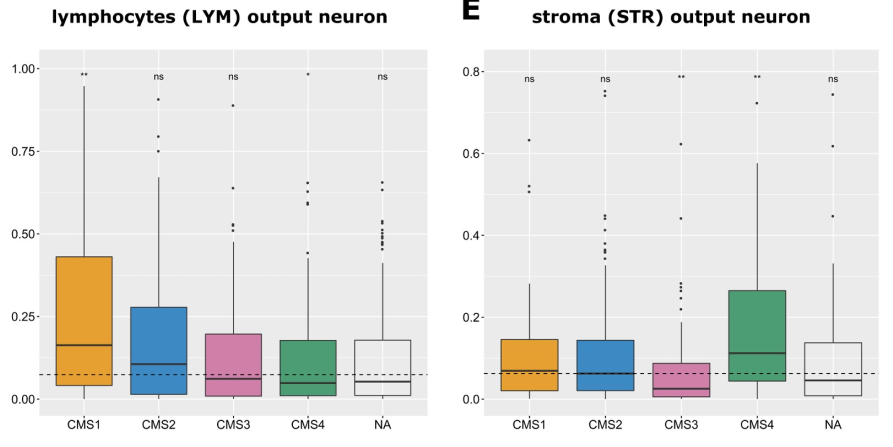
CMS1 tumor에서 Lymphocyte output neuron의 activation이 유의미하게 증가했고, CMS4 tumor에서 stroma output neuron의 activation이 유의미하게 높았음.
→ CNNS can decompose complex tissue parts and consistently identify issue components that are known to be present in specific molecular subtypes of CRC.
Deep stroma score and Cox analysis
Univariate Cox ph model을 통해 9개 중 5개의 class가 poor outcome과 correlate 된 것을 확인함.
adipose tissue: HR = 1.150 [n.s.]; debris: HR = 5.967 [p = 0.004]; lymphocytes: HR = 1.226 [n.s.]; muscle: HR = 3.761 [p = 0.025]; stroma: HR = 1.154 [n.s.]
이 5개의 class 정보를 활용하여 patient 별로 deep stroma score를 계산함.
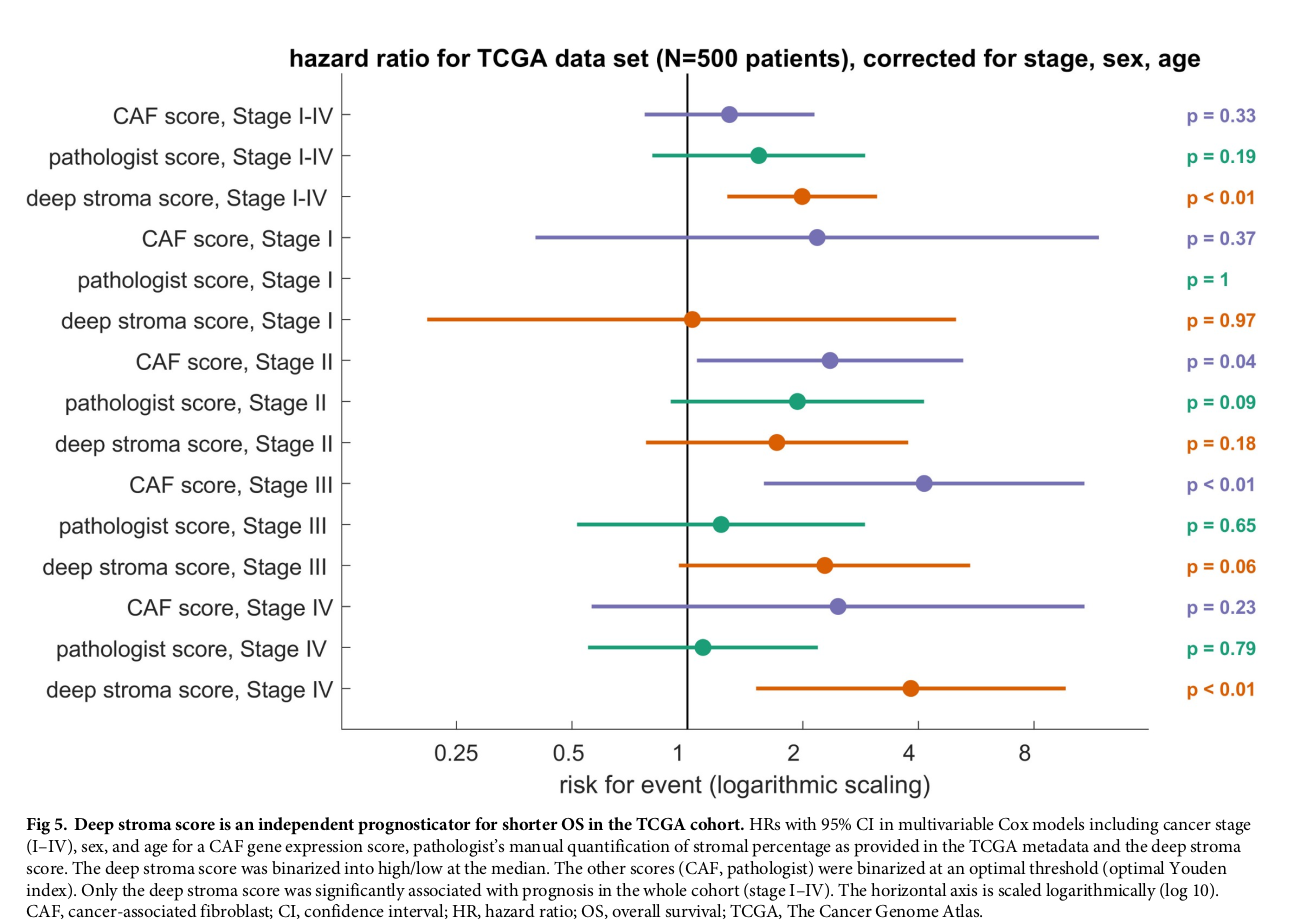
TCGA data set의 univariate / multivariate cox analysis를 prognostic factor임을 확인함.
(HR 2.12 [1.38–3.23], p = 0.001, HR 1.99 [1.27–3.12], p = 0.0028)

가정: There might be tumor-stage-specific differences in the prognostic power
Stage 별 deep stroma score 의 hazard ratio가 stage와 함께 증가함.
Stromal compartment의 gold standard method인 CAF score과 pathologist score과 비교함.
Full cohort에서 deep stroma score 만 prognostic power를 가짐.
CAF는 stage II, III에서, pathologic annotation은 없었고, DSS는 stage IV에서 power를 가짐.
CAF: stroma, extracellular matrix에서 발견되는 cancer-associated factor. CAF는 fibroblast를 quantify하고, pathologist는 이것 이외에 desmoplastic area도 quantify 함.
Desmoplastic stroma: growth of dense connect tissue occurs as result of injury or neoplasma
deep stroma score value 는 CAF score나 pathologist annotation과 상관관계를 보이지 않음. (p<0.05)
하지만 stroma component of deep stroma score는 CAF score과 상관관계를 보임. (person’s correlation coefficient is 0.26, p < 0.001). 이는 pathologist annotation / CAF score과의 상관관계 보다 높았음. (PCC is 0.20 < 0.001). → suggesting neural network is at least as good as pathologists at detecting the stromal component as reflected in gene expression analysis
Deep stroma generalization in different cohort
TCGA 와 동일한 cutoff value를 사용하여 DACHS study의 409 CRC patient에 대해 수행함.
OS 이외에도 diseases-specific survival (DSS), relapse-free survival (RFS) 고려함.
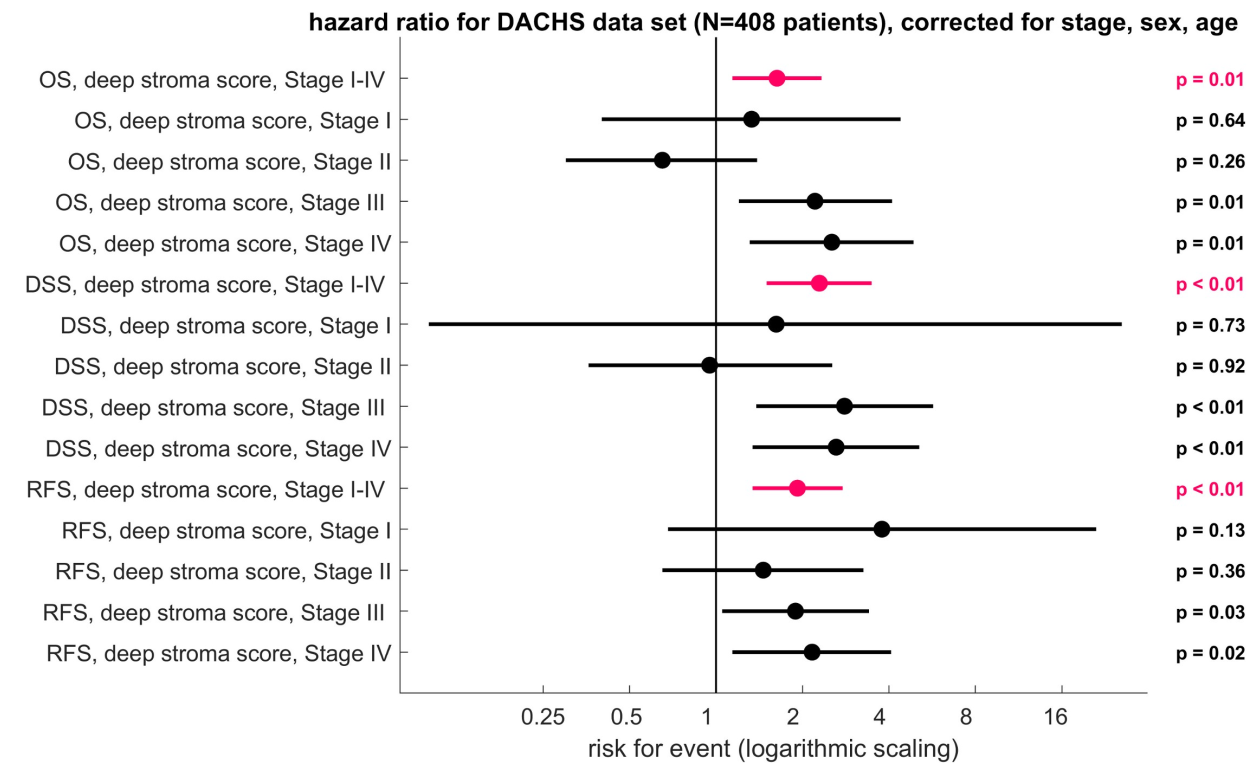
TCGA cohort의 결과와 마찬가지로, full cohort에 대해서 high significance를 보임.
OS (HR 1.63 [1.14–2.33], p = 0.008), DSS (HR 2.29 [1.5–3.48], p = 0.0004), and RFS (HR 1.92 [1.34–2.76], p = 0.0004) - this was independent of CRC stage, sex, age
또한 stage III와 IV에서도 disease specific survival (DSS) 결과 high significance를 보임.
(multivariable-adjusted CRC-specific survival for UICC stage 3: HR 2.8 [p = 0.0044]; stage
4: HR 2.62 [p = 0.0047])
Discussion
Deep stroma score significantly extends the UICC TNM system
이전 연구에서 stage II CRC를 image analysis로 보고했는데 우린 stage III, IV 함.
Stromal compartment에서 deep learning tech를 통해 prognostic information을 뽑아냄.
사람과 달리 CNN을 이용해 quantification of mixtures of different tissue를 수행함.
ex) 이 tile은 30% resemblance to desmoplastic stroma but 70% tumor epithelium
Conclusion
Tissue segmentation: tissue nine-component classification을 통해, pronostic power를 가진 feature의 HR weighted sum으로 deep stroma score를 계산하여 예후 예측에 활용함.
재밌..다!
