이 내용은 프로젝트를 진행하면서 배웠던 내용을 적용시켰지만, 점수가 떨어지는 현상이 발생되어서 추후에 원인을 다시 찾기 위한 코드입니다!
데이터셋 불러오기
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import koreanize_matplotlib
from sklearn.model_selection import train_test_split
from sklearn.tree import plot_tree
from sklearn.metrics import accuracy_score
train = pd.read_csv("와인품질분류/data/train.csv",index_col='index')
test = pd.read_csv("와인품질분류/data/test.csv",index_col='index')
submission = pd.read_csv('와인품질분류/data/sample_submission.csv')
submission.shape
(1000, 2)
print(train.shape, test.shape)
train.head(2)
(5497, 14) (1000, 12)
|
quality |
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
type |
qual_grade |
| index |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 0 |
5 |
5.6 |
0.695 |
0.06 |
6.8 |
0.042 |
9.0 |
84.0 |
0.99432 |
3.44 |
0.44 |
10.2 |
0 |
0.0 |
| 1 |
5 |
8.8 |
0.610 |
0.14 |
2.4 |
0.067 |
10.0 |
42.0 |
0.99690 |
3.19 |
0.59 |
9.5 |
1 |
0.0 |
test.head(2)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
type |
| index |
|
|
|
|
|
|
|
|
|
|
|
|
| 0 |
9.0 |
0.31 |
0.48 |
6.6 |
0.043 |
11.0 |
73.0 |
0.9938 |
2.90 |
0.38 |
11.6 |
white |
| 1 |
13.3 |
0.43 |
0.58 |
1.9 |
0.070 |
15.0 |
40.0 |
1.0004 |
3.06 |
0.49 |
9.0 |
red |
와인 성분 데이터 설명
- index 구분자
- quality 품질
- fixed acidity 산도
- volatile acidity 휘발성산
- citric acid 시트르산
- residual sugar 잔당 : 발효 후 와인 속에 남아있는 당분
- chlorides 염화물
- free sulfur dioxide 독립 이산화황
- total sulfur dioxide 총 이산화황
- density 밀도
- pH 수소이온농도
- sulphates 황산염
- alcohol 도수
- type 종류
중복 칼럼 제거
set(train.columns) - set(test.columns)
{'quality'}
결측치 확인
print(train.isnull().sum().sum(),test.isnull().sum().sum())
0 0
train.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 5497 entries, 0 to 5496
Data columns (total 13 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 quality 5497 non-null int64
1 fixed acidity 5497 non-null float64
2 volatile acidity 5497 non-null float64
3 citric acid 5497 non-null float64
4 residual sugar 5497 non-null float64
5 chlorides 5497 non-null float64
6 free sulfur dioxide 5497 non-null float64
7 total sulfur dioxide 5497 non-null float64
8 density 5497 non-null float64
9 pH 5497 non-null float64
10 sulphates 5497 non-null float64
11 alcohol 5497 non-null float64
12 type 5497 non-null object
dtypes: float64(11), int64(1), object(1)
memory usage: 601.2+ KB
test.info()
<class 'pandas.core.frame.DataFrame'>
Int64Index: 1000 entries, 0 to 999
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 fixed acidity 1000 non-null float64
1 volatile acidity 1000 non-null float64
2 citric acid 1000 non-null float64
3 residual sugar 1000 non-null float64
4 chlorides 1000 non-null float64
5 free sulfur dioxide 1000 non-null float64
6 total sulfur dioxide 1000 non-null float64
7 density 1000 non-null float64
8 pH 1000 non-null float64
9 sulphates 1000 non-null float64
10 alcohol 1000 non-null float64
11 type 1000 non-null object
dtypes: float64(11), object(1)
memory usage: 101.6+ KB
train.describe()
|
quality |
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| count |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
5497.000000 |
| mean |
5.818992 |
7.210115 |
0.338163 |
0.318543 |
5.438075 |
0.055808 |
30.417682 |
115.566491 |
0.994673 |
3.219502 |
0.530524 |
10.504918 |
| std |
0.870311 |
1.287579 |
0.163224 |
0.145104 |
4.756676 |
0.034653 |
17.673881 |
56.288223 |
0.003014 |
0.160713 |
0.149396 |
1.194524 |
| min |
3.000000 |
3.800000 |
0.080000 |
0.000000 |
0.600000 |
0.009000 |
1.000000 |
6.000000 |
0.987110 |
2.740000 |
0.220000 |
8.000000 |
| 25% |
5.000000 |
6.400000 |
0.230000 |
0.250000 |
1.800000 |
0.038000 |
17.000000 |
78.000000 |
0.992300 |
3.110000 |
0.430000 |
9.500000 |
| 50% |
6.000000 |
7.000000 |
0.290000 |
0.310000 |
3.000000 |
0.047000 |
29.000000 |
118.000000 |
0.994800 |
3.210000 |
0.510000 |
10.300000 |
| 75% |
6.000000 |
7.700000 |
0.400000 |
0.390000 |
8.100000 |
0.064000 |
41.000000 |
155.000000 |
0.996930 |
3.320000 |
0.600000 |
11.300000 |
| max |
9.000000 |
15.900000 |
1.580000 |
1.660000 |
65.800000 |
0.610000 |
289.000000 |
440.000000 |
1.038980 |
4.010000 |
2.000000 |
14.900000 |
quality 개수
train["quality"].value_counts().sort_index()
3 26
4 186
5 1788
6 2416
7 924
8 152
9 5
Name: quality, dtype: int64
sns.displot(train["quality"], kind='kde')
<seaborn.axisgrid.FacetGrid at 0x1fe85dd5df0>
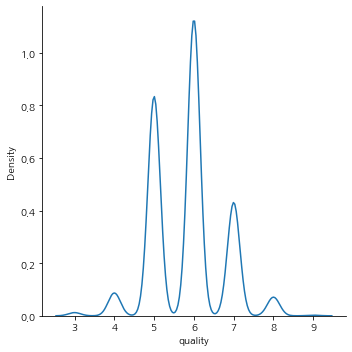
quality 척도 결정
train.loc[train['quality'] <= 6, 'qual_grade'] = 0
train.loc[train['quality'] > 6, 'qual_grade'] = 1
train['qual_grade'].value_counts(), train['qual_grade'].value_counts(normalize=True)
(0.0 4416
1.0 1081
Name: qual_grade, dtype: int64,
0.0 0.803347
1.0 0.196653
Name: qual_grade, dtype: float64)
데이터 전처리
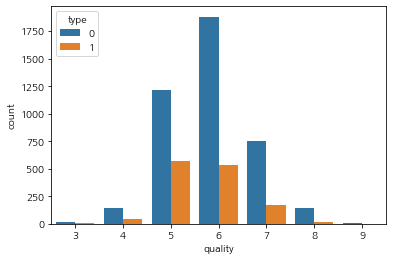
train['type'].value_counts(normalize=True), test['type'].value_counts(normalize=True)
(white 0.756595
red 0.243405
Name: type, dtype: float64,
white 0.739
red 0.261
Name: type, dtype: float64)
train["type"] = train["type"].map({"white":0,"red":1}).astype(int)
test["type"] = test["type"].map({"white":0,"red":1}).astype(int)
train['type'].value_counts(),test['type'].value_counts()
(0 4159
1 1338
Name: type, dtype: int64,
0 739
1 261
Name: type, dtype: int64)
sns.countplot(data= train, x= 'quality',hue="type")
<AxesSubplot:xlabel='quality', ylabel='count'>
상관관계
plt.figure(figsize=(10,6))
sns.heatmap(train.corr(), annot=True)
<AxesSubplot:>

plt.figure(figsize=(10,6))
sns.pairplot(data=train)
<seaborn.axisgrid.PairGrid at 0x1fe8d872fa0>
<Figure size 720x432 with 0 Axes>
train.corr()['quality'].sort_values()
density -0.299831
volatile acidity -0.261557
chlorides -0.198148
type -0.118280
fixed acidity -0.076506
total sulfur dioxide -0.039732
residual sugar -0.032848
pH 0.017931
sulphates 0.042068
free sulfur dioxide 0.055574
citric acid 0.079157
alcohol 0.439615
qual_grade 0.756656
quality 1.000000
Name: quality, dtype: float64
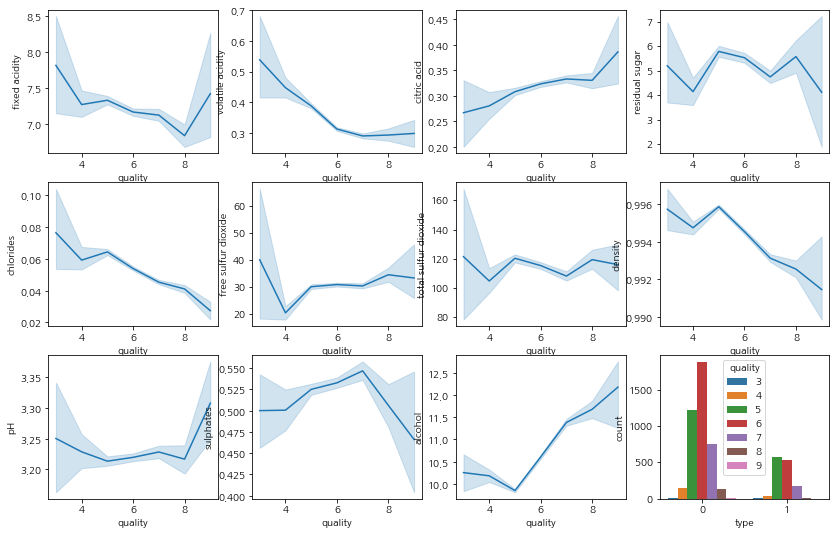
figure, ax = plt.subplots(3, 4)
figure.set_size_inches(14,9)
sns.lineplot(data=train, y="fixed acidity", x="quality", ax=ax[0][0])
sns.lineplot(data=train, y="volatile acidity", x="quality", ax=ax[0][1])
sns.lineplot(data=train, y="citric acid", x="quality", ax=ax[0][2])
sns.lineplot(data=train, y="residual sugar", x="quality", ax=ax[0][3])
sns.lineplot(data=train, y="chlorides", x="quality", ax=ax[1][0])
sns.lineplot(data=train, y="free sulfur dioxide", x="quality", ax=ax[1][1])
sns.lineplot(data=train, y="total sulfur dioxide", x="quality", ax=ax[1][2])
sns.lineplot(data=train, y="density", x="quality", ax=ax[1][3])
sns.lineplot(data=train, y="pH", x="quality", ax=ax[2][0])
sns.lineplot(data=train, y="sulphates", x="quality", ax=ax[2][1])
sns.lineplot(data=train, y="alcohol", x="quality", ax=ax[2][2])
sns.countplot(data=train, x='type', hue='quality', ax=ax[2][3])
<AxesSubplot:xlabel='type', ylabel='count'>
스케일링
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
s_scaler = StandardScaler()
m_scaler = MinMaxScaler()
train.columns
Index(['quality', 'fixed acidity', 'volatile acidity', 'citric acid',
'residual sugar', 'chlorides', 'free sulfur dioxide',
'total sulfur dioxide', 'density', 'pH', 'sulphates', 'alcohol', 'type',
'qual_grade'],
dtype='object')
s_col = ['fixed acidity', 'volatile acidity', 'citric acid', 'residual sugar',
'chlorides', 'free sulfur dioxide', 'total sulfur dioxide', 'density',
'pH', 'sulphates', 'alcohol']
print(train[s_col].shape)
train[s_col].head(5)
(5497, 11)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| index |
|
|
|
|
|
|
|
|
|
|
|
| 0 |
5.6 |
0.695 |
0.06 |
6.8 |
0.042 |
9.0 |
84.0 |
0.99432 |
3.44 |
0.44 |
10.2 |
| 1 |
8.8 |
0.610 |
0.14 |
2.4 |
0.067 |
10.0 |
42.0 |
0.99690 |
3.19 |
0.59 |
9.5 |
| 2 |
7.9 |
0.210 |
0.39 |
2.0 |
0.057 |
21.0 |
138.0 |
0.99176 |
3.05 |
0.52 |
10.9 |
| 3 |
7.0 |
0.210 |
0.31 |
6.0 |
0.046 |
29.0 |
108.0 |
0.99390 |
3.26 |
0.50 |
10.8 |
| 4 |
7.8 |
0.400 |
0.26 |
9.5 |
0.059 |
32.0 |
178.0 |
0.99550 |
3.04 |
0.43 |
10.9 |
train_sscale = s_scaler.fit_transform(train[s_col])
df_train = pd.DataFrame(train_sscale, columns=s_col)
train_mscale = m_scaler.fit_transform(train[s_col])
df_train = pd.DataFrame(train_mscale, columns=s_col)
df_train = pd.concat([df_train, train[s_col].reset_index()["index"]], axis=1)
df_train.set_index('index', inplace=True)
df_train.head(3)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| index |
|
|
|
|
|
|
|
|
|
|
|
| 0 |
-1.250611 |
2.186377 |
-1.78194 |
0.286345 |
-0.398500 |
-1.211937 |
-0.560852 |
-0.117252 |
1.372128 |
-0.605988 |
-0.255287 |
| 1 |
1.234899 |
1.665574 |
-1.23056 |
-0.638755 |
0.322998 |
-1.155351 |
-1.307080 |
0.738864 |
-0.183584 |
0.398147 |
-0.841348 |
| 2 |
0.535849 |
-0.785265 |
0.49250 |
-0.722855 |
0.034399 |
-0.532907 |
0.398583 |
-0.966732 |
-1.054782 |
-0.070450 |
0.330774 |
df_train.sample(10)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| 4782 |
0.247934 |
0.160000 |
0.234940 |
0.138037 |
0.028286 |
0.114583 |
0.271889 |
0.110854 |
0.346457 |
0.073034 |
0.594203 |
| 1632 |
0.363636 |
0.180000 |
0.240964 |
0.087423 |
0.049917 |
0.118056 |
0.359447 |
0.125120 |
0.322835 |
0.067416 |
0.565217 |
| 4942 |
0.214876 |
0.120000 |
0.126506 |
0.099693 |
0.051581 |
0.118056 |
0.359447 |
0.163678 |
0.511811 |
0.202247 |
0.275362 |
| 2786 |
0.181818 |
0.273333 |
0.000000 |
0.026074 |
0.098170 |
0.048611 |
0.062212 |
0.112011 |
0.661417 |
0.207865 |
0.652174 |
| 1165 |
0.305785 |
0.120000 |
0.150602 |
0.016871 |
0.048253 |
0.097222 |
0.283410 |
0.115867 |
0.559055 |
0.191011 |
0.347826 |
| 387 |
0.256198 |
0.160000 |
0.156627 |
0.026074 |
0.034942 |
0.034722 |
0.223502 |
0.076152 |
0.251969 |
0.112360 |
0.449275 |
| 5293 |
0.314050 |
0.173333 |
0.234940 |
0.107362 |
0.051581 |
0.152778 |
0.481567 |
0.181029 |
0.291339 |
0.174157 |
0.173913 |
| 2993 |
0.223140 |
0.166667 |
0.168675 |
0.084356 |
0.014975 |
0.138889 |
0.223502 |
0.079237 |
0.393701 |
0.056180 |
0.608696 |
| 51 |
0.380165 |
0.186667 |
0.216867 |
0.161043 |
0.038270 |
0.069444 |
0.290323 |
0.116059 |
0.165354 |
0.095506 |
0.724638 |
| 30 |
0.247934 |
0.240000 |
0.120482 |
0.236196 |
0.093178 |
0.208333 |
0.414747 |
0.226142 |
0.307087 |
0.129213 |
0.086957 |
test_sscale = s_scaler.fit_transform(test[s_col])
df_test = pd.DataFrame(test_sscale, columns=s_col)
test_mscale = m_scaler.fit_transform(test[s_col])
df_test = pd.DataFrame(test_mscale, columns=s_col)
df_test = pd.concat([df_test, test[s_col].reset_index()["index"]], axis=1)
df_test.set_index('index', inplace=True)
df_test.head(3)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| index |
|
|
|
|
|
|
|
|
|
|
|
| 0 |
1.306951 |
-0.220579 |
1.098184 |
0.236865 |
-0.385503 |
-1.108531 |
-0.756734 |
-0.352060 |
-1.943080 |
-1.068114 |
1.000172 |
| 1 |
4.507070 |
0.477272 |
1.780836 |
-0.749722 |
0.343645 |
-0.888114 |
-1.327873 |
1.915724 |
-0.949812 |
-0.311854 |
-1.203025 |
| 2 |
-0.553583 |
-0.395042 |
-0.335387 |
-0.057012 |
-0.466520 |
0.709907 |
1.077835 |
-0.008456 |
-0.142782 |
1.063164 |
-0.864071 |
모델링
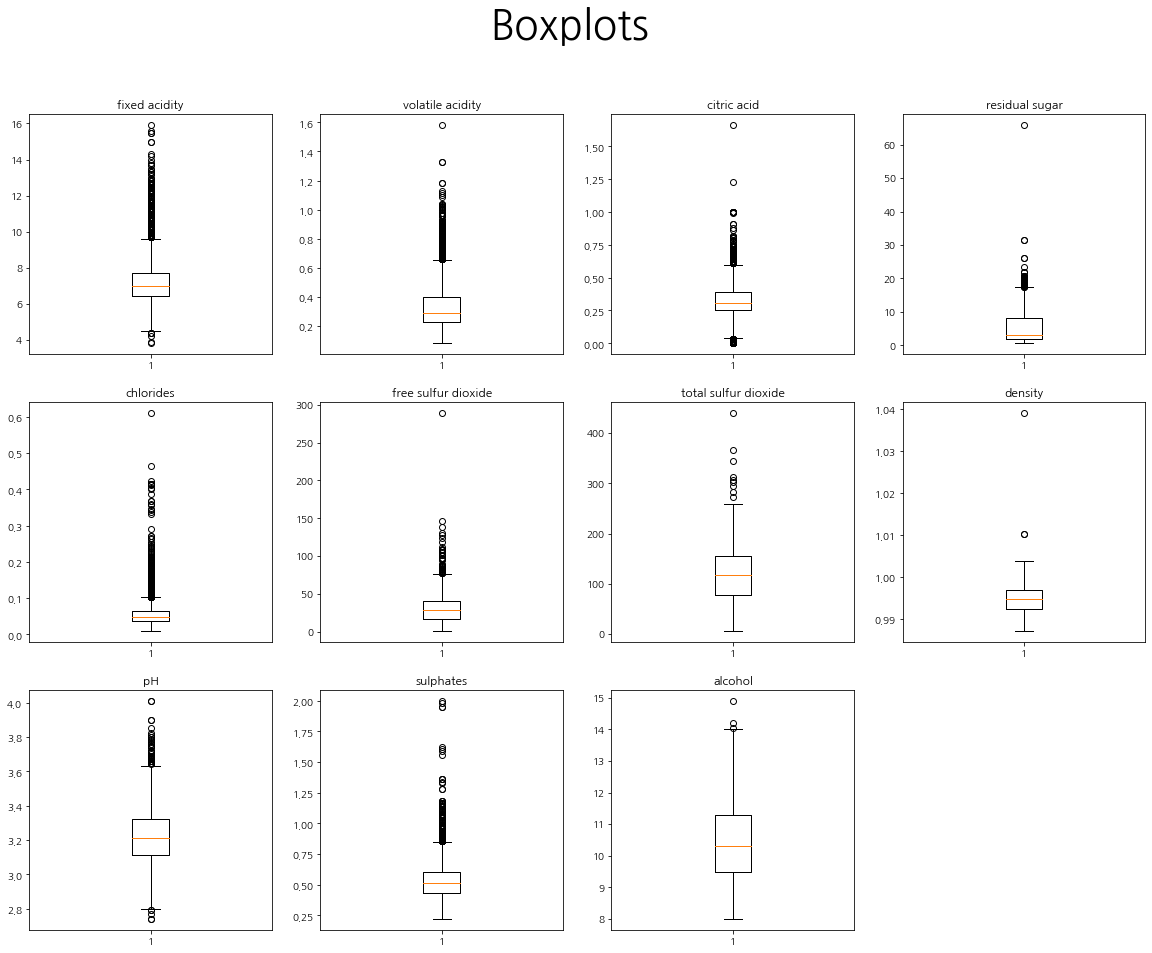
plt.figure(figsize=(20,15))
plt.suptitle("Boxplots", fontsize=40)
cols = train.columns[1:-2]
for i in range(len(cols)):
plt.subplot(3,4,i+1)
plt.title(cols[i])
plt.boxplot(train[cols[i]])
plt.show()
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import GradientBoostingClassifier
X_train = train.drop(columns=['type', 'quality', 'qual_grade'])
y_train = train['quality']
X_test = test.drop(columns=['type'])
X_train.head(1)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
| index |
|
|
|
|
|
|
|
|
|
|
|
| 0 |
5.6 |
0.695 |
0.06 |
6.8 |
0.042 |
9.0 |
84.0 |
0.99432 |
3.44 |
0.44 |
10.2 |
test.head(1)
|
fixed acidity |
volatile acidity |
citric acid |
residual sugar |
chlorides |
free sulfur dioxide |
total sulfur dioxide |
density |
pH |
sulphates |
alcohol |
type |
| index |
|
|
|
|
|
|
|
|
|
|
|
|
| 0 |
9.0 |
0.31 |
0.48 |
6.6 |
0.043 |
11.0 |
73.0 |
0.9938 |
2.9 |
0.38 |
11.6 |
0 |
df_train.columns
Index(['fixed acidity', 'volatile acidity', 'citric acid', 'residual sugar',
'chlorides', 'free sulfur dioxide', 'total sulfur dioxide', 'density',
'pH', 'sulphates', 'alcohol'],
dtype='object')
label_name = 'quality'
label_name
'quality'
feature_names=['fixed acidity',
'volatile acidity', 'citric acid',
'residual sugar',
'chlorides', 'free sulfur dioxide', 'total sulfur dioxide', 'density', 'pH',
'sulphates',
'alcohol']
feature_names
['fixed acidity',
'volatile acidity',
'citric acid',
'residual sugar',
'chlorides',
'free sulfur dioxide',
'total sulfur dioxide',
'density',
'pH',
'sulphates',
'alcohol']
X_train = df_train[feature_names]
y_train = train[label_name]
X_train.shape, y_train.shape
((5497, 11), (5497,))
X_test = df_test[feature_names]
X_test.shape
(1000, 11)
랜덤 포레스트
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=400, max_depth=12,min_samples_split=3
, min_samples_leaf=2,max_leaf_nodes=640, n_jobs=-1
,random_state=30)
model
RandomForestClassifier(max_depth=12, max_leaf_nodes=640, min_samples_leaf=2,
min_samples_split=3, n_estimators=400, n_jobs=-1,
random_state=30)
model.fit(X_train, y_train)
RandomForestClassifier(max_depth=12, max_leaf_nodes=640, min_samples_leaf=2,
min_samples_split=3, n_estimators=400, n_jobs=-1,
random_state=30)
y_predict = model.predict(X_test)
y_predict[:5]
array([6, 6, 6, 6, 6], dtype=int64)
submission['quality'] = y_predict
submission.head(2)
|
index |
quality |
| 0 |
0 |
6 |
| 1 |
1 |
6 |
submission.to_csv('baseline6.csv', index=False)
Validation (Evaluation)
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
def ACCURACY(true, pred):
score = np.mean(true==pred)
return score
X_fit, X_val, y_fit, y_val = train_test_split(X_train, y_train, test_size=0.3, stratify=y_train,
random_state=30)
model.fit(X_fit, y_fit)
RandomForestClassifier(max_depth=12, max_leaf_nodes=640, min_samples_leaf=2,
min_samples_split=3, n_estimators=400, n_jobs=-1,
random_state=30)
pred_val = model.predict(X_val)
accuracy_score(y_val, pred_val)
0.6424242424242425
print(classification_report(y_val, pred_val))
precision recall f1-score support
3 0.00 0.00 0.00 8
4 0.50 0.02 0.03 56
5 0.70 0.69 0.70 537
6 0.61 0.77 0.68 725
7 0.63 0.45 0.53 277
8 1.00 0.11 0.20 46
9 0.00 0.00 0.00 1
accuracy 0.64 1650
macro avg 0.49 0.29 0.31 1650
weighted avg 0.65 0.64 0.62 1650
C:\Users\dudal\anaconda3\lib\site-packages\sklearn\metrics\_classification.py:1318: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
C:\Users\dudal\anaconda3\lib\site-packages\sklearn\metrics\_classification.py:1318: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
C:\Users\dudal\anaconda3\lib\site-packages\sklearn\metrics\_classification.py:1318: UndefinedMetricWarning: Precision and F-score are ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
pd.DataFrame(model.feature_importances_,
index=X_train.columns,
columns=["f_Importances"]).sort_values(by="f_Importances", ascending=False)
|
f_Importances |
| alcohol |
0.152805 |
| density |
0.107966 |
| volatile acidity |
0.105885 |
| total sulfur dioxide |
0.086082 |
| chlorides |
0.085215 |
| sulphates |
0.083444 |
| residual sugar |
0.081187 |
| free sulfur dioxide |
0.079432 |
| pH |
0.077980 |
| citric acid |
0.073983 |
| fixed acidity |
0.066020 |
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import roc_auc_score, roc_curve, auc
import matplotlib.pyplot as plt
params = {
}
rf_clf_grid = RandomForestClassifier(random_state=30)
grid_rf_clf = GridSearchCV(estimator=rf_clf_grid, param_grid=params, cv=5,
scoring='accuracy', verbose=1, n_jobs=-1)
grid_rf_clf.fit(X_train, y_train)
Fitting 5 folds for each of 1 candidates, totalling 5 fits
GridSearchCV(cv=5, estimator=RandomForestClassifier(random_state=30), n_jobs=-1,
param_grid={}, scoring='accuracy', verbose=1)
print(grid_rf_clf.best_score_)
print(grid_rf_clf.best_estimator_)
0.6743618165274217
RandomForestClassifier(random_state=30)
best_model = grid_rf_clf.best_estimator_
best_model.fit(X_train,y_train)
RandomForestClassifier(random_state=30)
y_best_predict = best_model.predict(X_test)
y_best_predict
array([6, 6, 6, 5, 7, 6, 6, 6, 6, 6, 5, 5, 6, 6, 6, 5, 5, 6, 5, 5, 7, 6,
5, 6, 6, 6, 5, 5, 6, 5, 7, 6, 5, 5, 6, 6, 5, 5, 6, 5, 6, 6, 6, 6,
6, 6, 6, 6, 6, 5, 6, 5, 5, 6, 5, 5, 6, 6, 6, 6, 5, 5, 5, 6, 6, 5,
5, 5, 5, 5, 5, 5, 5, 5, 6, 5, 5, 6, 6, 6, 6, 6, 5, 5, 6, 5, 5, 5,
6, 6, 6, 6, 5, 6, 6, 6, 5, 6, 6, 6, 5, 5, 6, 5, 5, 6, 6, 6, 6, 6,
7, 6, 6, 5, 7, 6, 5, 6, 7, 5, 6, 5, 6, 7, 6, 6, 6, 6, 6, 6, 6, 5,
5, 5, 5, 6, 5, 6, 6, 5, 5, 6, 6, 6, 5, 5, 6, 5, 6, 6, 6, 7, 6, 5,
5, 5, 5, 6, 5, 5, 5, 5, 6, 6, 6, 6, 6, 6, 6, 6, 5, 6, 6, 5, 6, 6,
6, 6, 5, 6, 6, 5, 6, 6, 5, 6, 6, 6, 5, 6, 6, 6, 6, 6, 6, 6, 7, 6,
5, 6, 6, 6, 6, 6, 6, 5, 7, 6, 6, 5, 6, 5, 6, 6, 6, 6, 6, 6, 6, 6,
6, 6, 6, 6, 6, 6, 6, 5, 5, 5, 6, 7, 7, 6, 6, 6, 6, 6, 6, 6, 7, 6,
6, 6, 7, 6, 6, 7, 6, 7, 6, 5, 6, 7, 6, 6, 6, 7, 6, 6, 7, 6, 6, 6,
5, 7, 6, 6, 6, 6, 7, 5, 6, 5, 6, 6, 7, 6, 6, 5, 6, 6, 6, 6, 6, 6,
6, 6, 5, 5, 5, 5, 6, 6, 5, 5, 5, 5, 5, 7, 6, 5, 5, 5, 5, 6, 5, 6,
6, 5, 5, 6, 6, 5, 6, 6, 6, 5, 5, 6, 5, 5, 5, 5, 5, 6, 5, 6, 7, 5,
6, 6, 6, 6, 5, 6, 5, 6, 6, 5, 6, 6, 5, 6, 6, 7, 5, 7, 6, 6, 5, 5,
5, 6, 6, 5, 5, 6, 6, 6, 5, 6, 5, 6, 6, 6, 5, 5, 6, 6, 6, 5, 6, 6,
6, 5, 5, 5, 6, 5, 6, 6, 6, 7, 5, 6, 6, 5, 6, 5, 6, 5, 7, 5, 5, 6,
6, 6, 6, 6, 6, 6, 5, 6, 6, 6, 6, 6, 6, 6, 6, 6, 5, 5, 6, 6, 6, 5,
5, 6, 6, 6, 5, 6, 5, 6, 5, 6, 5, 6, 7, 5, 5, 6, 6, 5, 6, 5, 5, 5,
5, 6, 6, 6, 6, 6, 6, 5, 5, 6, 6, 6, 5, 6, 5, 5, 6, 6, 6, 5, 6, 6,
6, 5, 5, 5, 6, 6, 6, 5, 5, 6, 5, 6, 5, 6, 6, 6, 6, 6, 6, 6, 6, 5,
6, 6, 6, 7, 6, 5, 6, 6, 6, 6, 5, 6, 5, 5, 6, 5, 6, 6, 7, 6, 6, 6,
6, 6, 6, 6, 6, 6, 6, 6, 6, 5, 6, 6, 6, 6, 6, 6, 5, 6, 6, 5, 6, 6,
6, 6, 6, 6, 6, 6, 7, 6, 6, 6, 6, 6, 6, 5, 6, 6, 7, 6, 6, 5, 5, 6,
6, 6, 7, 6, 5, 5, 5, 6, 6, 6, 6, 5, 5, 6, 6, 7, 6, 6, 5, 6, 6, 5,
5, 6, 6, 6, 6, 6, 6, 5, 6, 6, 6, 5, 6, 7, 7, 6, 6, 6, 6, 5, 6, 6,
5, 5, 6, 6, 5, 6, 6, 7, 6, 6, 6, 6, 5, 5, 6, 6, 6, 5, 6, 5, 6, 6,
5, 6, 7, 6, 6, 6, 5, 6, 5, 6, 5, 6, 6, 6, 5, 6, 5, 5, 5, 6, 6, 6,
6, 5, 5, 6, 6, 5, 5, 6, 6, 6, 5, 5, 6, 5, 6, 6, 6, 6, 7, 6, 6, 5,
6, 6, 6, 6, 6, 6, 7, 5, 5, 5, 6, 5, 5, 7, 6, 5, 6, 6, 6, 6, 6, 6,
6, 5, 6, 7, 6, 6, 6, 7, 7, 6, 6, 6, 6, 5, 6, 6, 5, 5, 6, 6, 5, 6,
6, 6, 5, 7, 5, 5, 6, 5, 6, 6, 6, 6, 5, 6, 6, 7, 6, 6, 6, 6, 6, 6,
5, 6, 7, 6, 5, 6, 6, 6, 7, 6, 5, 6, 6, 5, 6, 6, 6, 5, 5, 6, 6, 5,
6, 5, 6, 5, 6, 6, 6, 7, 5, 6, 6, 6, 6, 6, 6, 8, 6, 6, 5, 5, 6, 6,
6, 6, 6, 5, 6, 6, 6, 6, 7, 5, 6, 5, 5, 5, 5, 6, 6, 5, 5, 6, 6, 6,
6, 6, 6, 6, 7, 5, 6, 6, 5, 5, 6, 6, 6, 6, 5, 5, 6, 6, 6, 6, 6, 6,
7, 5, 6, 6, 5, 6, 6, 6, 5, 6, 5, 6, 6, 5, 6, 6, 6, 7, 6, 6, 6, 6,
7, 7, 5, 6, 5, 5, 7, 5, 7, 6, 6, 6, 6, 6, 5, 6, 5, 6, 5, 6, 6, 5,
5, 5, 6, 6, 6, 5, 5, 6, 6, 6, 6, 6, 7, 6, 6, 6, 5, 6, 6, 6, 7, 6,
6, 6, 6, 5, 5, 7, 6, 6, 6, 5, 5, 6, 6, 6, 6, 6, 5, 7, 6, 6, 6, 5,
5, 7, 5, 5, 5, 6, 5, 6, 6, 5, 6, 6, 6, 6, 6, 7, 5, 5, 7, 6, 7, 6,
6, 6, 6, 5, 5, 6, 6, 6, 6, 6, 6, 6, 5, 6, 5, 6, 5, 6, 6, 5, 6, 6,
6, 6, 6, 6, 6, 6, 5, 5, 6, 6, 6, 7, 6, 6, 5, 6, 6, 5, 5, 6, 5, 6,
6, 6, 6, 6, 6, 6, 6, 6, 7, 6, 5, 6, 6, 6, 5, 5, 5, 6, 6, 5, 5, 5,
6, 6, 6, 6, 6, 5, 6, 6, 6, 6], dtype=int64)
submission['quality'] = y_best_predict
submission.to_csv('baseline7.csv', index=False)
cross validation 으로 학습 세트의 오차 측정하기
from sklearn.model_selection import cross_val_predict
y_pred = cross_val_predict(model, X_train, y_train, cv=5, n_jobs=-1, verbose=3)
y_pred[:5]
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers.
[Parallel(n_jobs=-1)]: Done 2 out of 5 | elapsed: 8.7s remaining: 13.0s
[Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 8.9s finished
array([5, 5, 6, 6, 6], dtype=int64)
(y_train == y_pred).mean()
0.5517555030016372
y_train.shape[0]
실제값과 예측값의 차이 시각화 하기
sns.regplot(x=y_train, y=y_predict)
sns.jointplot(x=y_train, y=y_predict)
from sklearn.metrics import r2_score
r2_score(y_train, y_predict)
df_y = pd.DataFrame({"true" : y_train, "predict" : y_predict})
df_y.shape
df_y.plot(kind="kde")
sns.displot(df_y, kde=True, aspect= 5)
오차구하기
error = abs(y_train - y_predict)
error
error.describe()
sns.displot(error, aspect=5)
MAE
mae = abs(y_train - y_predict).mean()
mae
MAPE
mape = (abs(y_train - y_predict)/y_train).mean()
mape
MSE
mse = ((y_train - y_predict) ** 2).mean()
mse
RMSE
rmse = np.sqrt(mse)
rmse
GridSearchCV
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import roc_auc_score, roc_curve, auc
import matplotlib.pyplot as plt
max_depth = list(range(3,20,2))
max_depth
[3, 5, 7, 9, 11, 13, 15, 17, 19]
max_features = [0.3, 0.5, 0.7, 0.8, 0.9 ]
max_features
[0.3, 0.5, 0.7, 0.8, 0.9]
params = {'n_estimators' : [100, 150, 200,500,1000],
'criterion' : ['gini', 'entropy']}
clf = GridSearchCV(model, param_grid=params, cv=5, return_train_score=True, verbose=2)
clf.fit(X_train,y_train)
y_pred = clf.predict(X_test)
Fitting 5 folds for each of 10 candidates, totalling 50 fits
[CV] END ...................criterion=gini, n_estimators=100; total time= 0.0s
[CV] END ...................criterion=gini, n_estimators=100; total time= 0.0s
[CV] END ...................criterion=gini, n_estimators=100; total time= 0.0s
[CV] END ...................criterion=gini, n_estimators=100; total time= 0.0s
[CV] END ...................criterion=gini, n_estimators=100; total time= 0.0s
[CV] END ...................criterion=gini, n_estimators=150; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=150; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=150; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=150; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=150; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=200; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=200; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=200; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=200; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=200; total time= 0.1s
[CV] END ...................criterion=gini, n_estimators=500; total time= 0.4s
[CV] END ...................criterion=gini, n_estimators=500; total time= 0.4s
[CV] END ...................criterion=gini, n_estimators=500; total time= 0.3s
[CV] END ...................criterion=gini, n_estimators=500; total time= 0.3s
[CV] END ...................criterion=gini, n_estimators=500; total time= 0.3s
[CV] END ..................criterion=gini, n_estimators=1000; total time= 0.8s
[CV] END ..................criterion=gini, n_estimators=1000; total time= 0.7s
[CV] END ..................criterion=gini, n_estimators=1000; total time= 0.8s
[CV] END ..................criterion=gini, n_estimators=1000; total time= 0.8s
[CV] END ..................criterion=gini, n_estimators=1000; total time= 0.7s
[CV] END ................criterion=entropy, n_estimators=100; total time= 0.0s
[CV] END ................criterion=entropy, n_estimators=100; total time= 0.0s
[CV] END ................criterion=entropy, n_estimators=100; total time= 0.0s
[CV] END ................criterion=entropy, n_estimators=100; total time= 0.0s
[CV] END ................criterion=entropy, n_estimators=100; total time= 0.0s
[CV] END ................criterion=entropy, n_estimators=150; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=150; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=150; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=150; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=150; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=200; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=200; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=200; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=200; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=200; total time= 0.1s
[CV] END ................criterion=entropy, n_estimators=500; total time= 0.4s
[CV] END ................criterion=entropy, n_estimators=500; total time= 0.4s
[CV] END ................criterion=entropy, n_estimators=500; total time= 0.4s
[CV] END ................criterion=entropy, n_estimators=500; total time= 0.4s
[CV] END ................criterion=entropy, n_estimators=500; total time= 0.4s
[CV] END ...............criterion=entropy, n_estimators=1000; total time= 0.8s
[CV] END ...............criterion=entropy, n_estimators=1000; total time= 0.8s
[CV] END ...............criterion=entropy, n_estimators=1000; total time= 0.8s
[CV] END ...............criterion=entropy, n_estimators=1000; total time= 1.1s
[CV] END ...............criterion=entropy, n_estimators=1000; total time= 0.9s
clf.best_estimator_
RandomForestClassifier(max_depth=5, min_samples_leaf=7, n_estimators=150,
n_jobs=-1, random_state=42)
pd.DataFrame(clf.cv_results_).sort_values("rank_test_score")
|
mean_fit_time |
std_fit_time |
mean_score_time |
std_score_time |
param_criterion |
param_n_estimators |
params |
split0_test_score |
split1_test_score |
split2_test_score |
... |
mean_test_score |
std_test_score |
rank_test_score |
split0_train_score |
split1_train_score |
split2_train_score |
split3_train_score |
split4_train_score |
mean_train_score |
std_train_score |
| 1 |
0.145958 |
0.009643 |
0.030632 |
0.001710 |
gini |
150 |
{'criterion': 'gini', 'n_estimators': 150} |
0.568182 |
0.560909 |
0.552320 |
... |
0.555573 |
0.007828 |
1 |
0.578576 |
0.586536 |
0.578445 |
0.582538 |
0.584584 |
0.582136 |
0.003219 |
| 2 |
0.169492 |
0.008166 |
0.039045 |
0.001134 |
gini |
200 |
{'criterion': 'gini', 'n_estimators': 200} |
0.568182 |
0.558182 |
0.554140 |
... |
0.555027 |
0.008400 |
2 |
0.578121 |
0.584262 |
0.582992 |
0.583902 |
0.582538 |
0.582363 |
0.002209 |
| 0 |
0.098874 |
0.002276 |
0.023209 |
0.000244 |
gini |
100 |
{'criterion': 'gini', 'n_estimators': 100} |
0.569091 |
0.558182 |
0.552320 |
... |
0.555027 |
0.008189 |
3 |
0.577439 |
0.587673 |
0.578445 |
0.587312 |
0.586176 |
0.583409 |
0.004502 |
| 3 |
0.358002 |
0.016511 |
0.089475 |
0.002779 |
gini |
500 |
{'criterion': 'gini', 'n_estimators': 500} |
0.564545 |
0.557273 |
0.549591 |
... |
0.553572 |
0.007172 |
4 |
0.577439 |
0.587446 |
0.580264 |
0.586630 |
0.580946 |
0.582545 |
0.003861 |
| 4 |
0.693886 |
0.023758 |
0.174820 |
0.003033 |
gini |
1000 |
{'criterion': 'gini', 'n_estimators': 1000} |
0.564545 |
0.557273 |
0.551410 |
... |
0.553208 |
0.008173 |
5 |
0.578121 |
0.584262 |
0.581173 |
0.586403 |
0.583902 |
0.582772 |
0.002859 |
| 8 |
0.404777 |
0.013810 |
0.089389 |
0.003571 |
entropy |
500 |
{'criterion': 'entropy', 'n_estimators': 500} |
0.563636 |
0.556364 |
0.554140 |
... |
0.553026 |
0.009175 |
6 |
0.571526 |
0.571526 |
0.569577 |
0.578217 |
0.571851 |
0.572539 |
0.002951 |
| 9 |
0.773880 |
0.004096 |
0.220118 |
0.093729 |
entropy |
1000 |
{'criterion': 'entropy', 'n_estimators': 1000} |
0.565455 |
0.554545 |
0.552320 |
... |
0.551389 |
0.010076 |
7 |
0.571753 |
0.571753 |
0.569804 |
0.578445 |
0.573215 |
0.572994 |
0.002933 |
| 5 |
0.102412 |
0.001495 |
0.023178 |
0.000297 |
entropy |
100 |
{'criterion': 'entropy', 'n_estimators': 100} |
0.560909 |
0.553636 |
0.553230 |
... |
0.550298 |
0.008579 |
8 |
0.572663 |
0.571981 |
0.565484 |
0.579582 |
0.572306 |
0.572403 |
0.004464 |
| 7 |
0.178822 |
0.002047 |
0.041150 |
0.001132 |
entropy |
200 |
{'criterion': 'entropy', 'n_estimators': 200} |
0.557273 |
0.551818 |
0.549591 |
... |
0.547933 |
0.009124 |
9 |
0.568797 |
0.571981 |
0.567303 |
0.578672 |
0.573897 |
0.572130 |
0.004009 |
| 6 |
0.148497 |
0.001769 |
0.030472 |
0.000999 |
entropy |
150 |
{'criterion': 'entropy', 'n_estimators': 150} |
0.558182 |
0.558182 |
0.543221 |
... |
0.547932 |
0.009764 |
10 |
0.570389 |
0.572663 |
0.567076 |
0.578217 |
0.572078 |
0.572085 |
0.003631 |
10 rows × 22 columns
RandomizedSearchCV
np.random.randint(3, 20, 10)
np.random.uniform(0.5, 1, 10)
from sklearn.model_selection import RandomizedSearchCV
param_distributions = {"max_depth" : np.random.randint(3, 20, 10),
"max_features" : np.random.uniform(0.5, 1, 10)}
clfr = RandomizedSearchCV(model,
param_distributions=param_distributions,
n_iter=10,
cv=5,
scoring="accuracy",
n_jobs=-1,
random_state=42, verbose=3)
clfr.fit(X_train,y_train)
clfr.best_estimator_
clfr.best_params_
pd.DataFrame(clfr.cv_results_).nsmallest(5,"rank_test_score")
Best Estimator
best_model = clfr.best_estimator_
best_model.fit(X_train, y_train)
y_predict = best_model.predict(X_test)
y_predict
모델평가하기
best_model.feature_importances_
sns.barplot(x=best_model.feature_importances_, y=best_model.feature_names_in_)
submission = pd.read_csv('와인품질분류/data/sample_submission.csv')
submission['quality'] = y_predict
submission
submission.to_csv('baseline3.csv', index=False)
X_train = train.drop(['index', 'quality'], axis = 1)
y_train = train['quality']
X_test = test.drop('index', axis = 1)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(train_x, train_y)
y_pred = model.predict(X_test)
from sklearn.model_selection import cross_val_predict, cross_validate, cross_val_score
y_valid_predict = cross_val_predict(model, X_train, y_train, cv=5, n_jobs=-1, verbose=3)
y_valid_predict
cross_val_score(model, X_train, y_train).mean()
submission = pd.read_csv('와인품질분류/data/sample_submission.csv')
submission['quality'] = y_pred
submission
submission.to_csv('baseline2.csv', index=False)
```python
train.head(2)
label_name = 'quality'
label_name
train.columns
feature_names = ['fixed acidity', 'volatile acidity', 'citric acid',
'residual sugar', 'chlorides', 'free sulfur dioxide',
'total sulfur dioxide', 'density', 'pH', 'sulphates', 'alcohol',
'type']
X_train = train[feature_names]
y_train = train[label_name]
X_test = test[feature_names]
print(set(X_train.columns) - set(X_test.columns))
print(set(X_test.columns) - set(X_train.columns))
X = train[feature_names]
y = train[label_name]
X.shape, y.shape
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, stratify=y, random_state=42)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
plt.figure(figsize=(5,3))
sns.countplot(x=y_train)
plt.figure(figsize=(5,3))
sns.countplot(x=y_test)
y_train.value_counts(normalize=True)
y_test.value_counts(normalize=True)
머신러닝 모델
from sklearn.tree import DecisionTreeClassifier
model = DecisionTreeClassifier(max_depth=10,max_features=0.8,random_state=42)
model
model.fit(X_train, y_train)
from sklearn.model_selection import cross_val_predict, cross_validate, cross_val_score
y_valid_predict = cross_val_predict(model, X_train, y_train, cv=5, n_jobs=-1, verbose=3)
y_valid_predict
pd.DataFrame(cross_validate(model, X_train, y_train, cv=5))
cross_val_score(model, X_train, y_train).mean()
(y_train == y_valid_predict).mean()
y_train.shape[0]
y_predict = model.predict(X_test)
print(y_predict.shape)
y_predict
pd.score
Grid SearchCV
max_depth = list(range(3,30,2))
max_depth
max_features = [0.3, 0.5, 0.7, 0.8, 0.9 ]
max_features
parameters = {"max_depth" : max_depth, "max_features" : max_features}
parameters
from sklearn.model_selection import GridSearchCV
clf = GridSearchCV(model, parameters, n_jobs=-1, cv=5)
clf.fit(X_train,y_train)
clf.best_estimator_
pd.DataFrame(clf.cv_results_).sort_values("rank_test_score")
Random
np.random.randint(3, 20, 10)
np.random.uniform(0.5, 1, 10)
from sklearn.model_selection import RandomizedSearchCV
param_distributions = {"max_depth" : np.random.randint(3, 20, 10),
"max_features" : np.random.uniform(0.5, 1, 10)}
clfr = RandomizedSearchCV(model,
param_distributions=param_distributions,
n_iter=10,
cv=5,
scoring="accuracy",
n_jobs=-1,
random_state=42, verbose=3)
clfr.fit(X_train,y_train)
clfr.best_estimator_
clfr.best_params_
pd.DataFrame(clfr.cv_results_).nsmallest(5,"rank_test_score")
submission = pd.read_csv('와인품질분류/data/sample_submission.csv')
submission['quality'] = y_predict
submission
submission.to_csv('baseline.csv', index=False)







그냥 읽고만 가기에는 너무 정리가 잘 된 글이네요!!
좋은 글 잘 읽고 갑니다!!!